Most recent BDB-Lab papers
BDB-Lab Links
Major Projects
Other Links
Lab Members
Twitter feed
Tweets by BigDataBiologyPublications
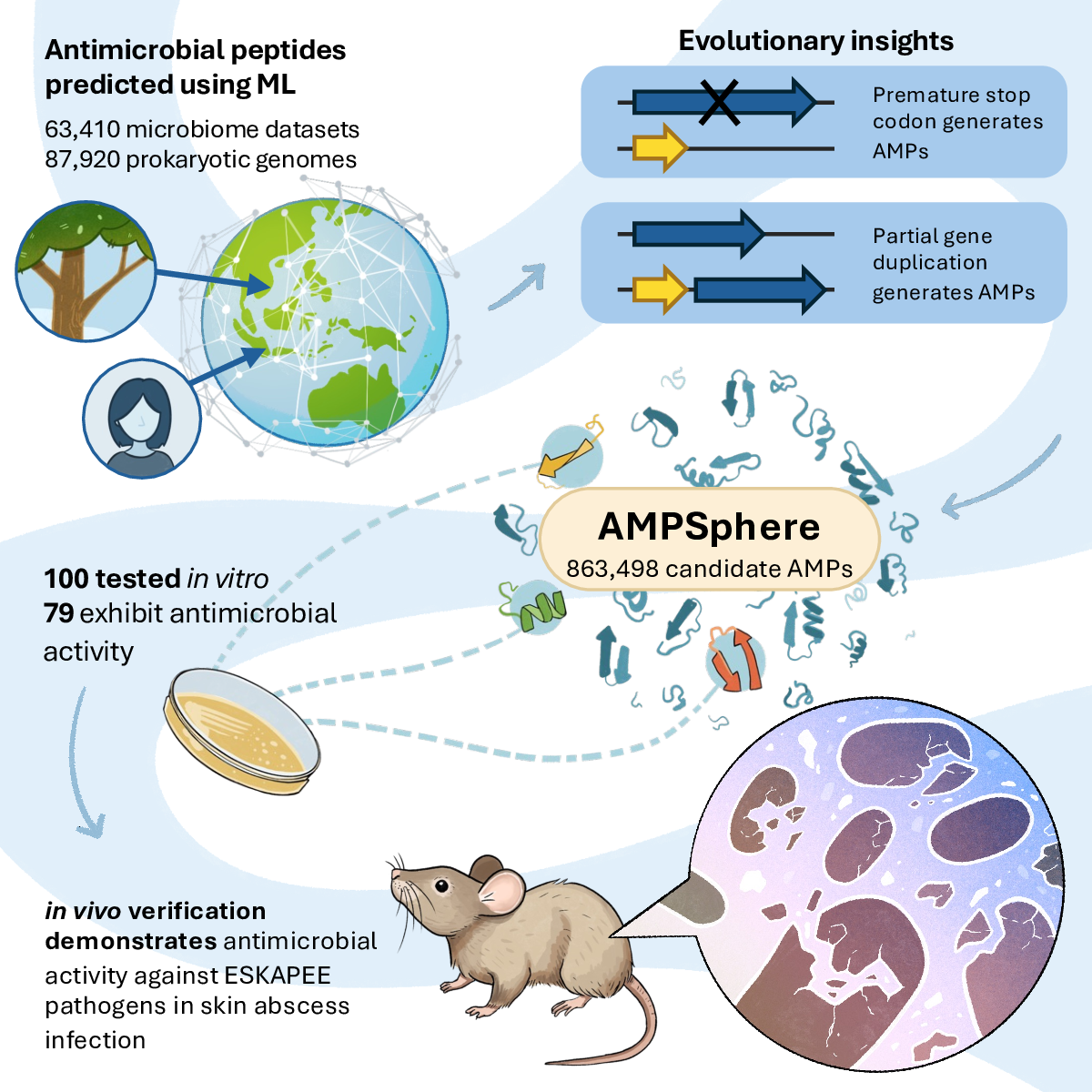
36. Discovery of antimicrobial peptides in the global microbiome with machine learning
by Célio Dias Santos Júnior, Marcelo D.T. Torres, Yiqian Duan, ..., Hui Chong, ..., Chengkai Zhu, Amy Houseman, ..., Cesar de la Fuente-Nunez, Luis Pedro Coelho in Cell
35. For long-term sustainable software in bioinformatics
by Luis Pedro Coelho in PLOS Computational Biology
34. Challenges in computational discovery of bioactive peptides in ’omics data
by Luis Pedro Coelho, Célio Dias Santos Júnior, Cesar de la Fuente‐Nunez in PROTEOMICS
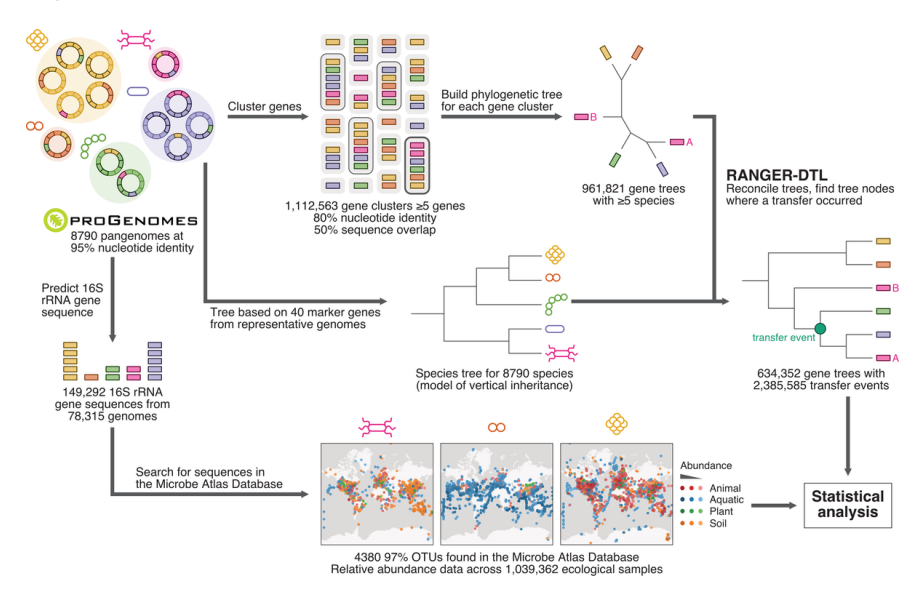
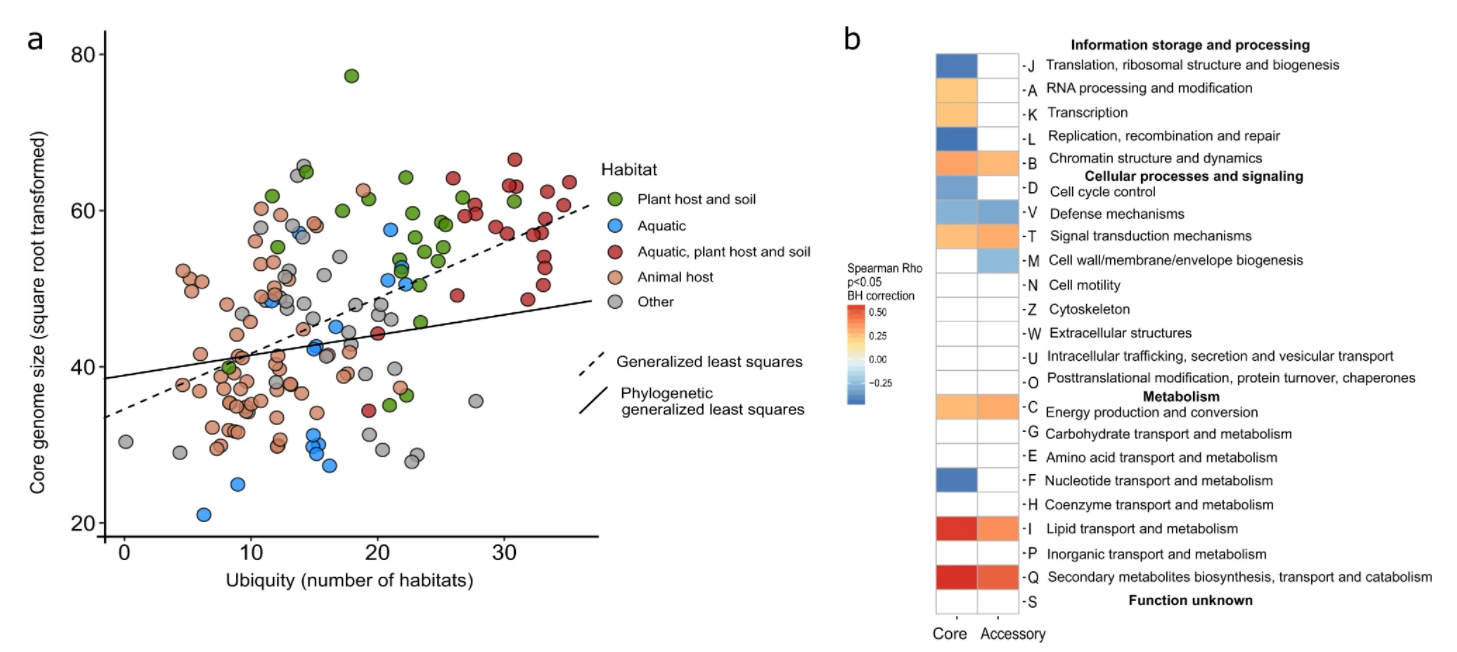
33. A global survey of prokaryotic genomes reveals the eco-evolutionary pressures driving horizontal gene transfer
by Marija Dmitrijeva, Janko Tackmann, João Frederico Matias Rodrigues, Jaime Huerta-Cepas, Luis Pedro Coelho, Christian von Mering in Nature Ecology & Evolution
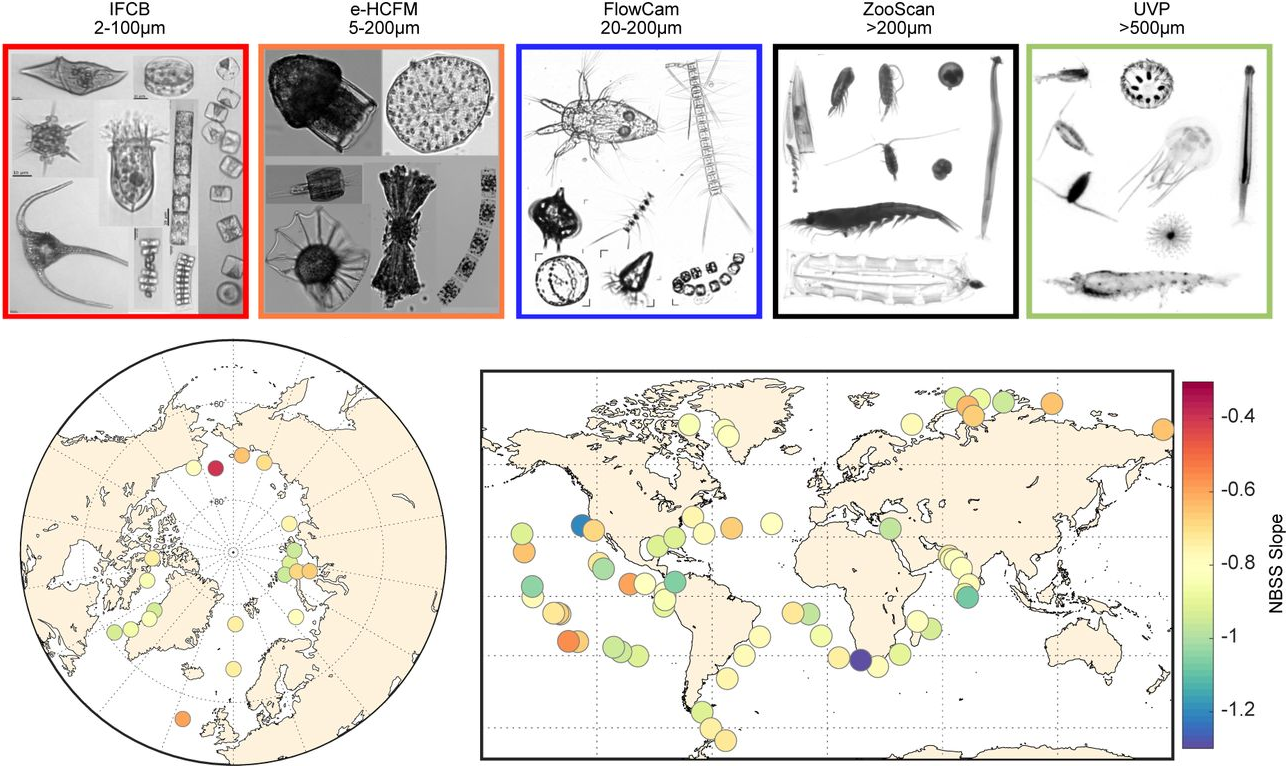
32. Ubiquity of inverted ’gelatinous’ ecosystem pyramids in the global ocean
by Fabien Lombard, Lionel Guidi, Manoela C. Brandão, Luis Pedro Coelho, ..., Gabriel Gorsky, Tara Oceans Coordinators in bioRxiv (PREPRINT)
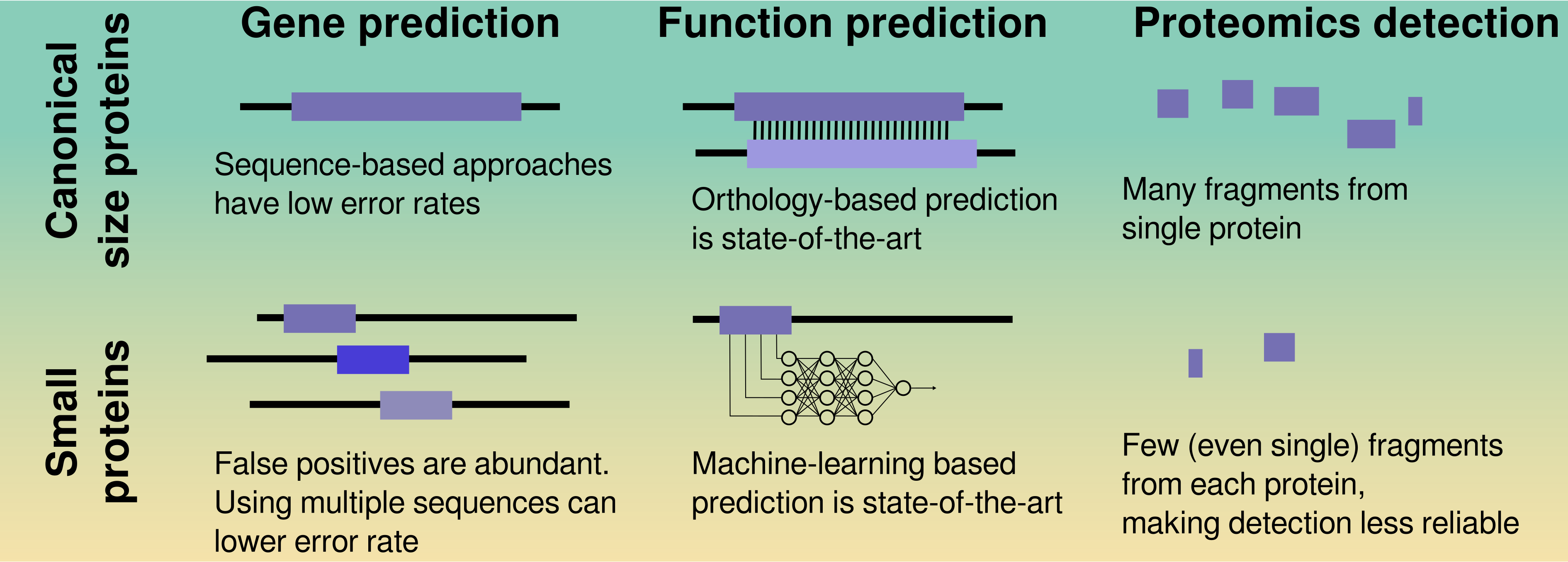
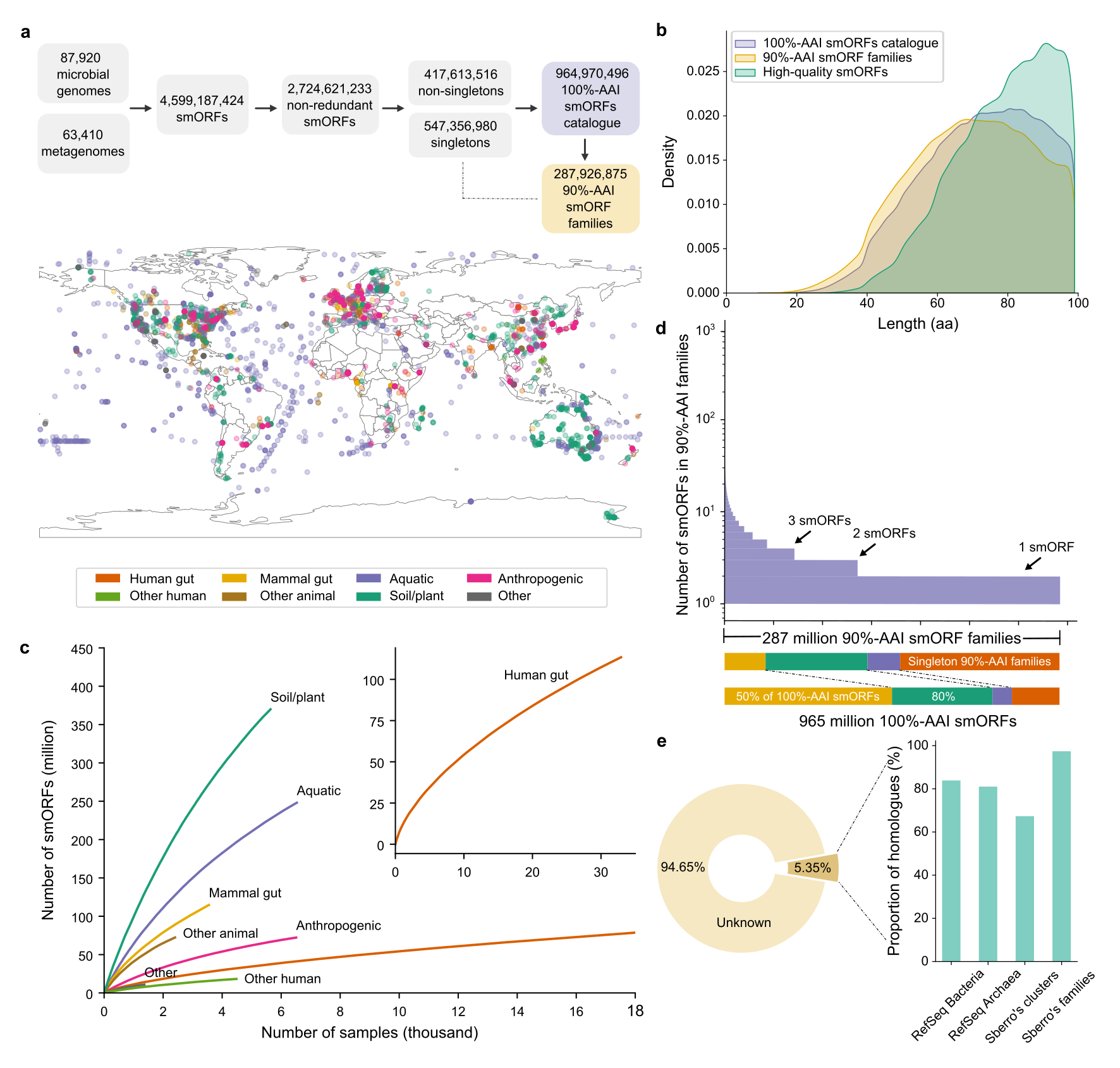
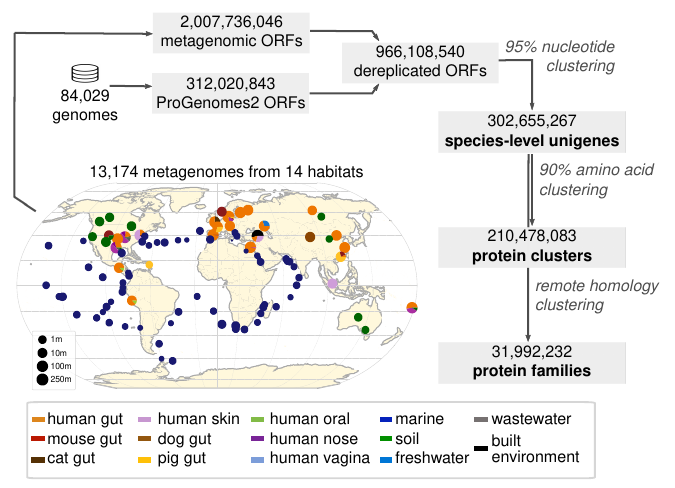
31. A catalogue of small proteins from the global microbiome
by Yiqian Duan, Célio Dias Santos Júnior, Thomas S.B. Schmidt, Anthony Fullam, Breno L. S. de Almeida, Chengkai Zhu, Kuhn Michael, Xing-Ming Zhao, Peer Bork, Luis Pedro Coelho in biorXiv (PREPRINT)
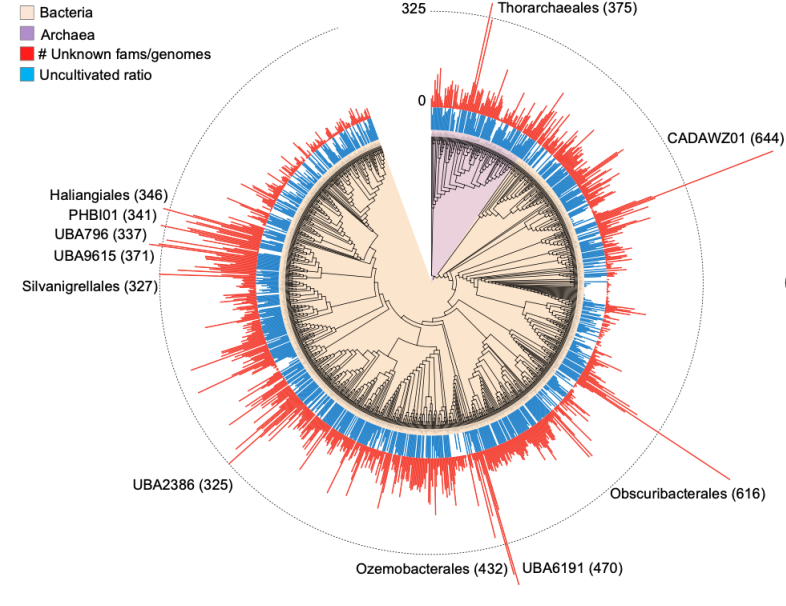
30. Functional and evolutionary significance of unknown genes from uncultivated taxa
by Álvaro Rodríguez del Río, Joaquín Giner-Lamia, Carlos P. Cantalapiedra, ..., Luis Pedro Coelho, Jaime Huerta-Cepas in Nature
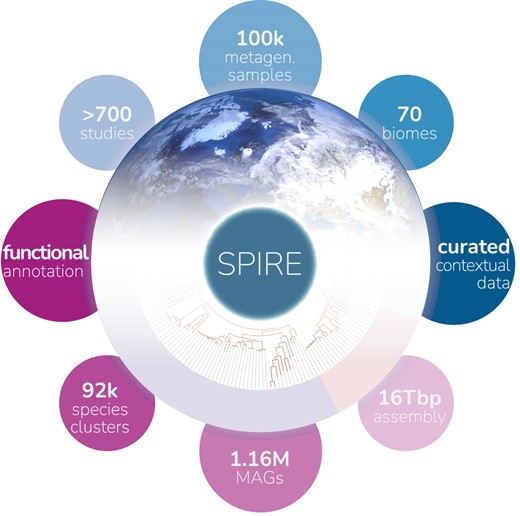
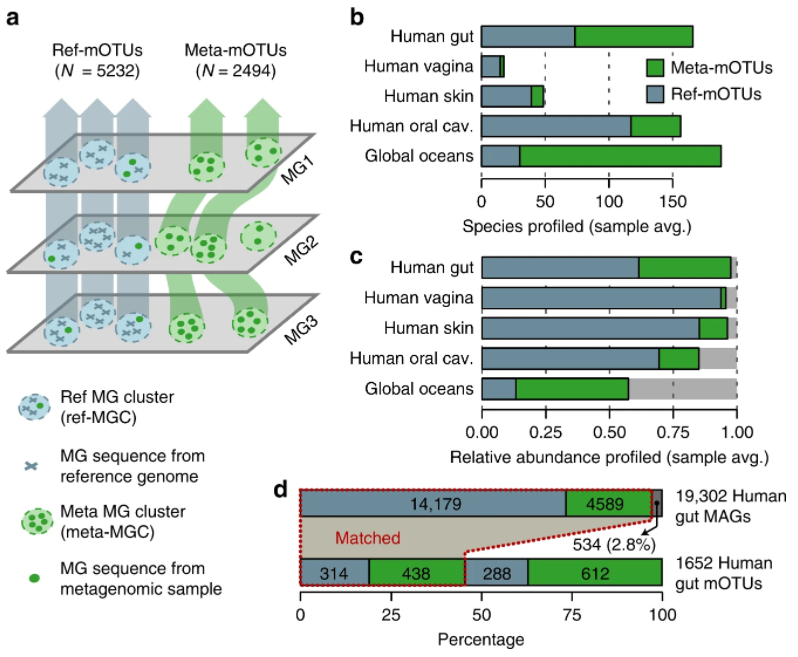
29. SPIRE: a Searchable, Planetary-scale mIcrobiome REsource
by Thomas S B Schmidt, Anthony Fullam, Pamela Ferretti, ..., Yiqian Duan, ..., Luis Pedro Coelho, Peer Bork in Nucleic Acids Research
DOI: 10.1093/nar/gkad943
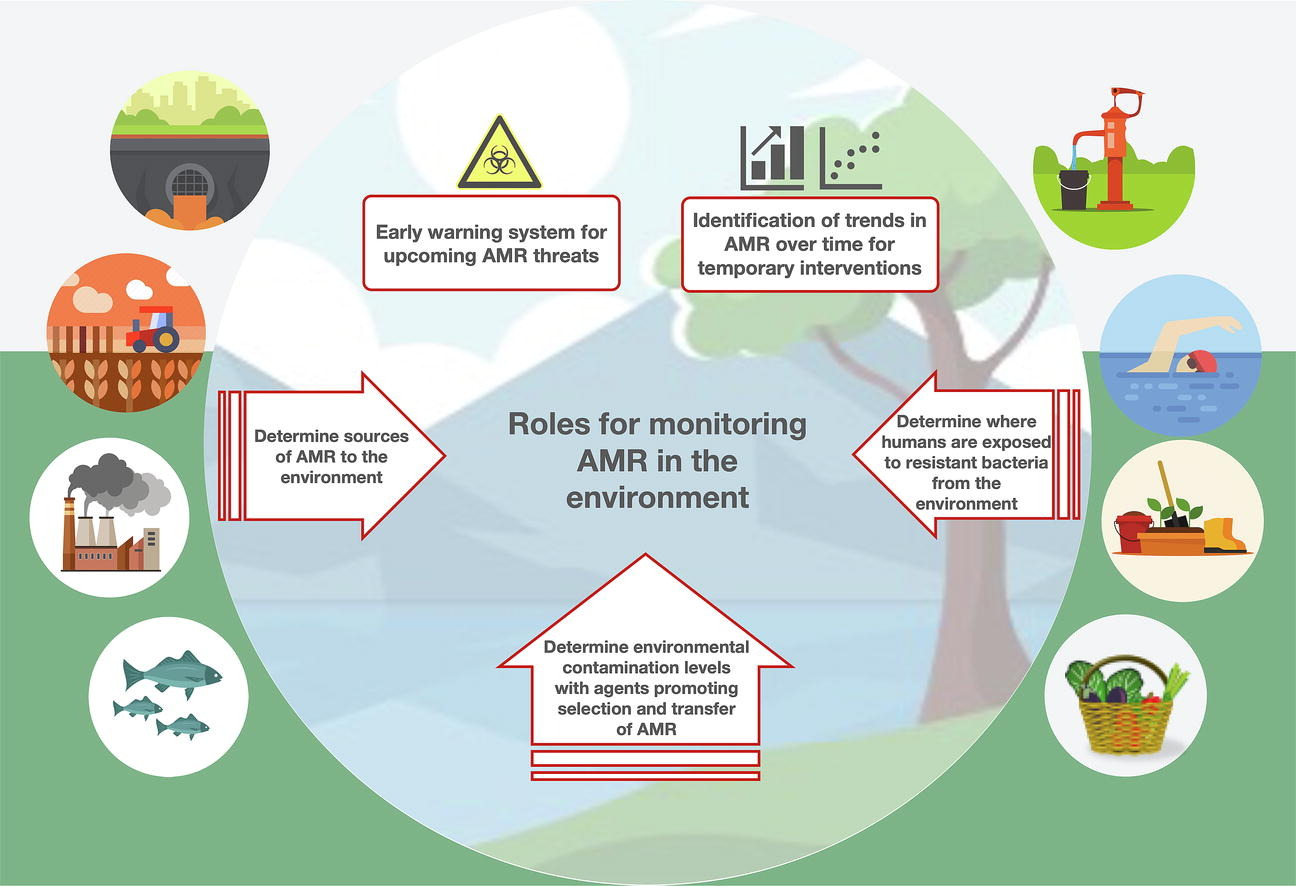
28. Towards Monitoring of Antimicrobial Resistance in the Environment: For what Reasons, How to Implement It, and What Are the Data Needs?
by Johan Bengtsson-Palme, Anna Abramova, Thomas U. Berendonk, Luis Pedro Coelho, ..., Svetlana Ugarcina Perovic, ..., Astrid Louise Wester, Rabaab Zahra in Environment International
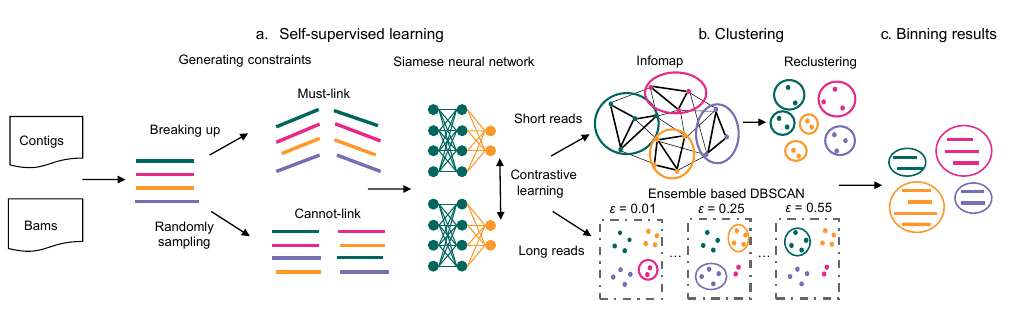
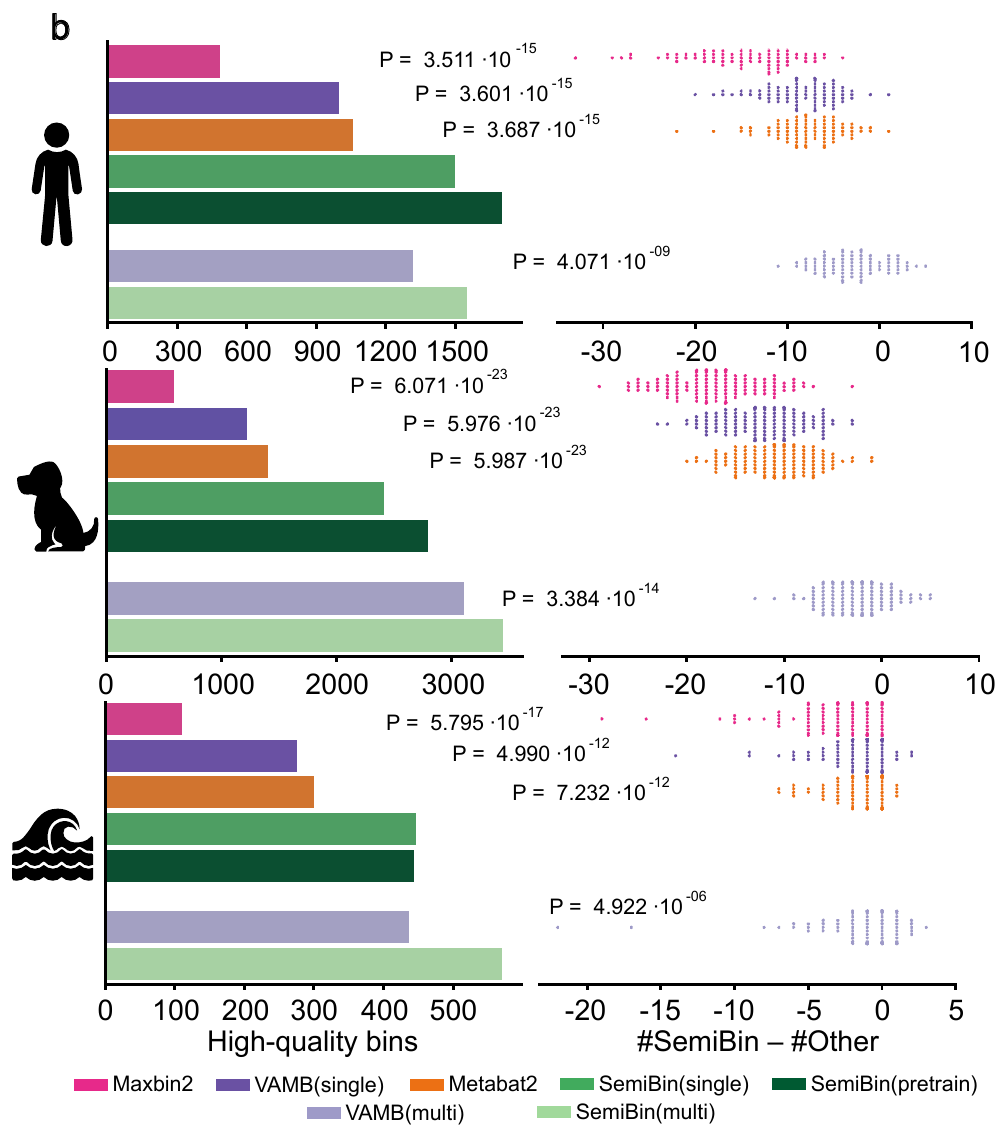
27. SemiBin2: self-supervised contrastive learning leads to better MAGs for short- and long-read sequencing
by Shaojun Pan, Xing-Ming Zhao, Luis Pedro Coelho in Bioinformatics
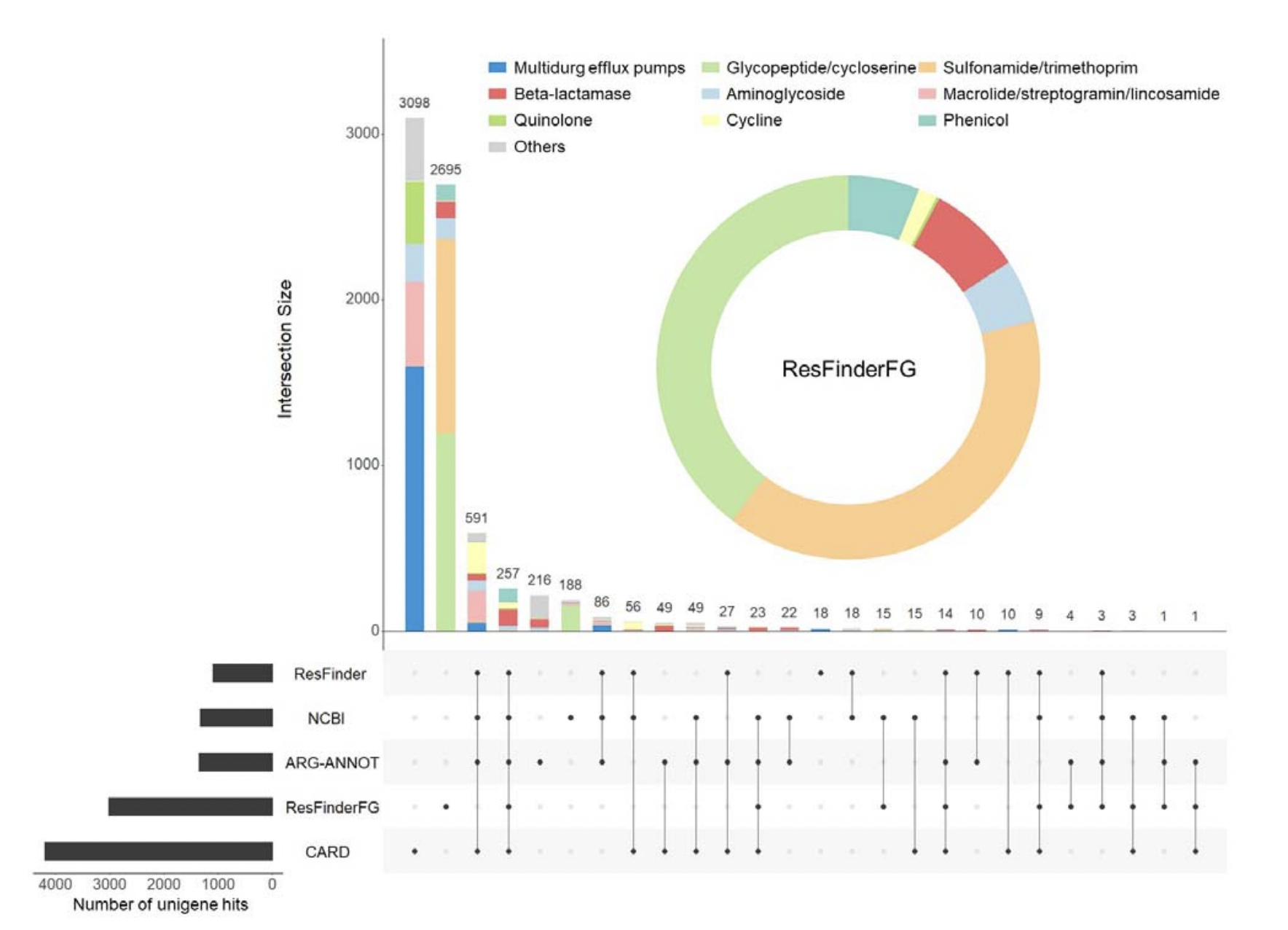
26. ResFinderFG v2.0: a database of antibiotic resistance genes obtained by functional metagenomics
by Rémi Gschwind, Svetlana Ugarcina Perovic, Maja Weiss, Marie Petitjean, Julie Lao, Luis Pedro Coelho, Etienne Ruppé in Nucleic Acids Research
DOI: 10.1093/nar/gkad384
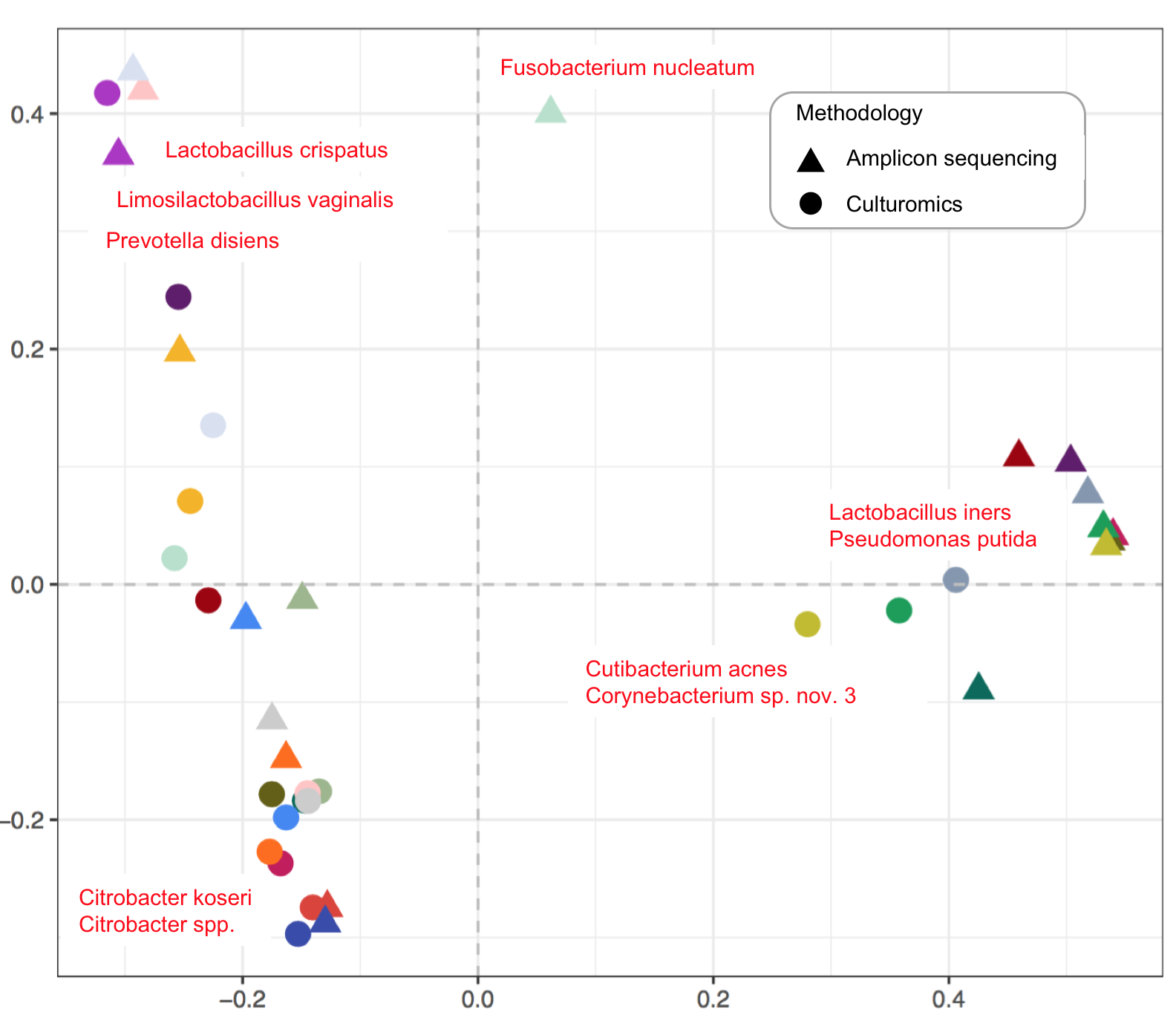
25. Urinary Microbiome of Reproductive-Age Asymptomatic European Women
by Svetlana Ugarcina Perovic, Magdalena Ksiezarek, Joana Rocha, Elisabete Alves Cappelli, Márcia Sousa, Teresa Gonçalves Ribeiro, Filipa Grosso, Luísa Peixe in Microbiology Spectrum
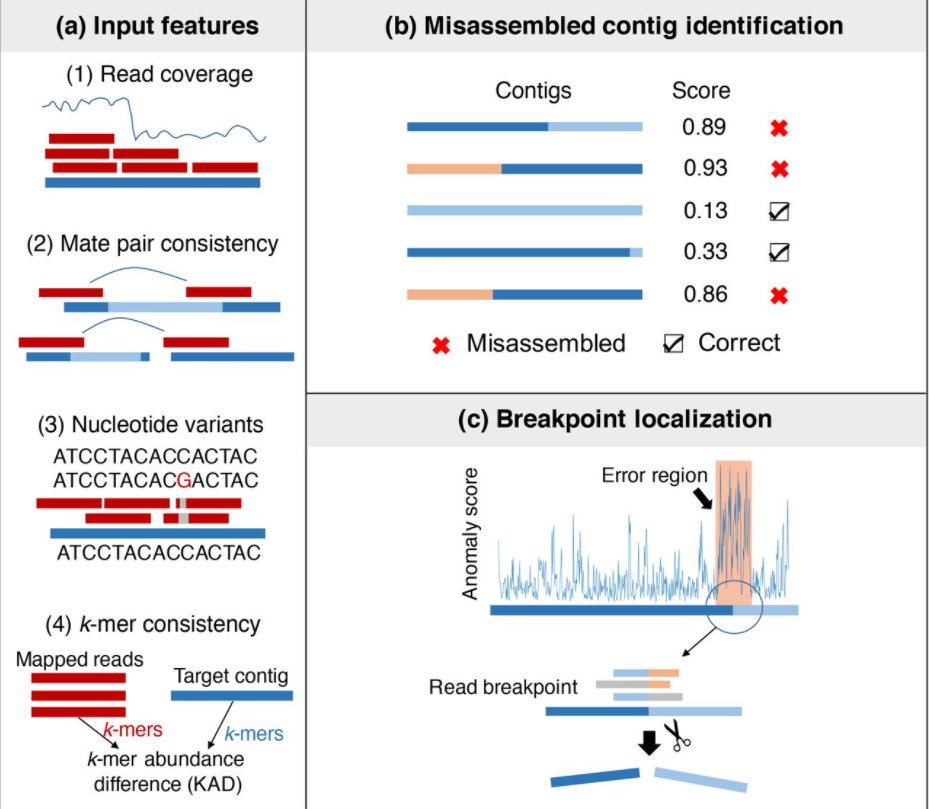
24. metaMIC: reference-free Misassembly Identification and Correction of de novo metagenomic assemblies
by Senying Lai, Shaojun Pan, Chuqing Sun, Luis Pedro Coelho, Wei-Hua Chen, Xing-Ming Zhao in Genome biology
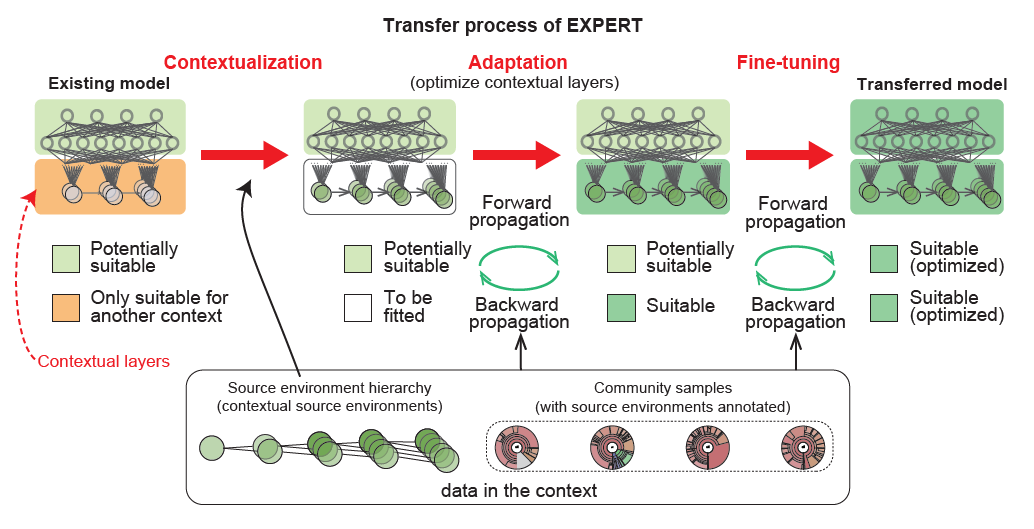
23. EXPERT: Transfer Learning-enabled context-aware microbial source tracking
by Hui Chong, Yuguo Zha, Qingyang Yu, ..., Luis Pedro Coelho, Kang Ning in Briefings in Bioinformatics
DOI: 10.1093/bib/bbac396
22. Drivers and Determinants of Strain Dynamics Following Faecal Microbiota Transplantation
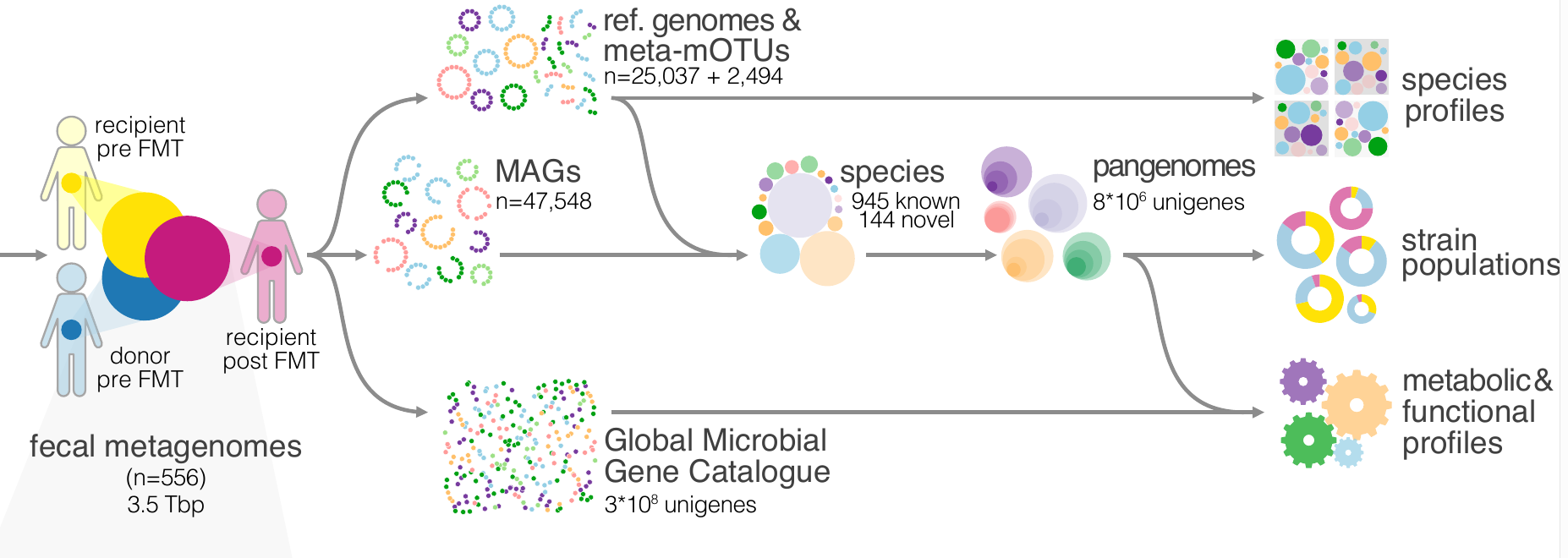
[More about Drivers and Determinants of Strain Dynamics Following Faecal Microbiota Transplantation]
by Thomas SB Schmidt, Simone S Li, Oleksandr M Maistrenko, ..., Luis Pedro Coelho, ..., Max Nieuwdorp, Peer Bork in Nature Medicine
21. A deep siamese neural network improves metagenome-assembled genomes in microbiome datasets across different environments
by Shaojun Pan, Chengkai Zhu, Xing-Ming Zhao, Luis Pedro Coelho in Nature Communications
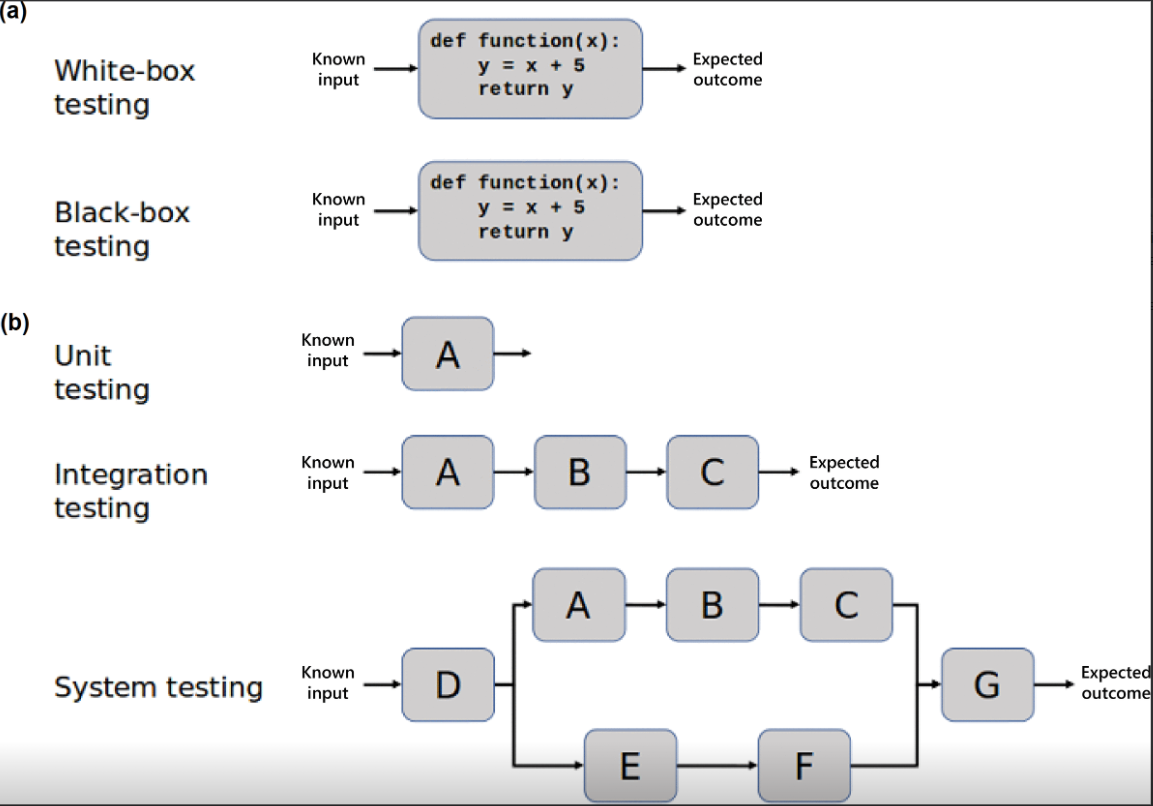
20. Software testing in microbial bioinformatics: a call to action
by Boas C.L. van der Putten, C. I. Mendes, Brooke M. Talbot, Jolinda de Korne-Elenbaas, Rafael Mamede, Pedro Vila-Cerqueira, Luis Pedro Coelho, Christopher A. Gulvik, Lee S. Katz, The ASM NGS 2020 Hackathon participants in Microbial Genomics
19. Microbiome and metabolome features of the cardiometabolic disease spectrum
by Sebastien Fromentin, Sofia K. Forslund, Kanta Chechi, ..., Luis Pedro Coelho, ..., S. Dusko Ehrlich, Oluf Pedersen in Nature Medicine
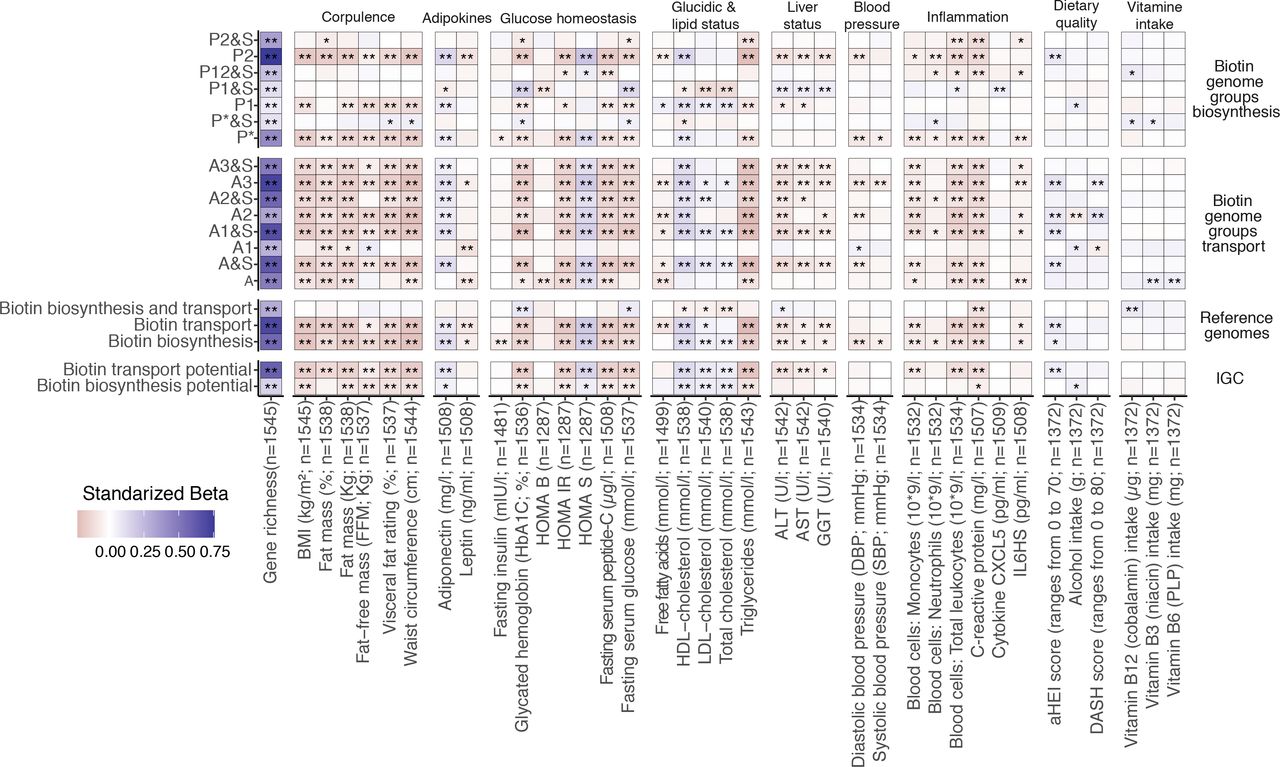
18. Impairment of gut microbial biotin metabolism and host biotin status in severe obesity: effect of biotin and prebiotic supplementation on improved metabolism
by Eugeni Belda, Lise Voland, Valentina Tremaroli, ..., Luis Pedro Coelho, ..., Karine Clément, The MetaCardis Consortium in Gut
17. Towards the biogeography of prokaryotic genes
by Luis Pedro Coelho, Renato Alves, Álvaro Rodríguez del Río, ..., Shaojun Pan, ..., Jaime Huerta-Cepas, Peer Bork in Nature
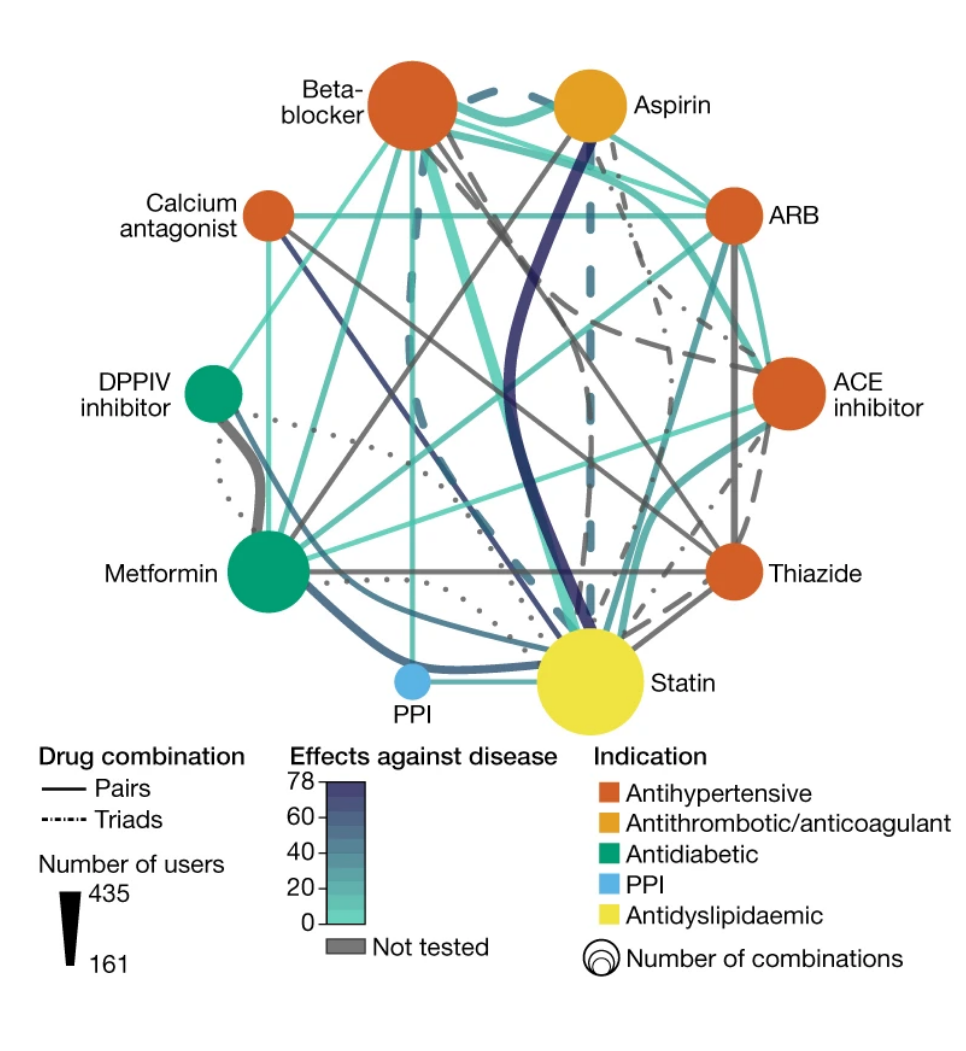
16. Combinatorial, additive and dose-dependent drug–microbiome associations
by Sofia K. Forslund, Rima Chakaroun, Maria Zimmermann-Kogadeeva, ..., Luis Pedro Coelho, ..., Peer Bork, The MetaCardis Consortium* in Nature
15. An open code pledge for the neuroscience community
by Cooper Smout, Dawn Liu Holford, Kelly Garner, ..., Sina Mansour L., Luis Pedro Coelho in Preprint
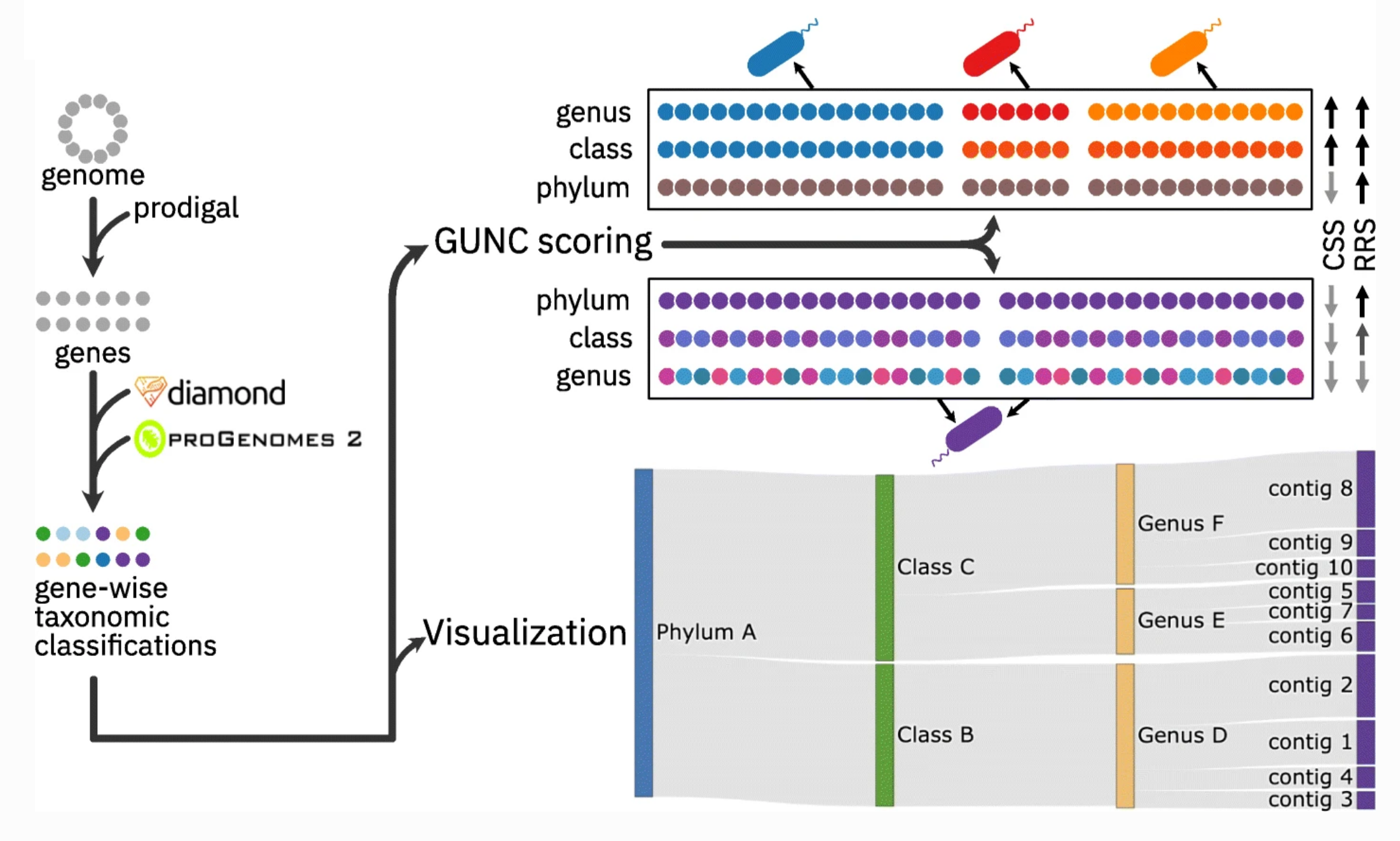
14. GUNC: detection of chimerism and contamination in prokaryotic genomes
by Askarbek Orakov, Anthony Fullam, Luis Pedro Coelho, Supriya Khedkar, Damian Szklarczyk, Daniel R. Mende, Thomas S. B. Schmidt, Peer Bork in Genome Biology
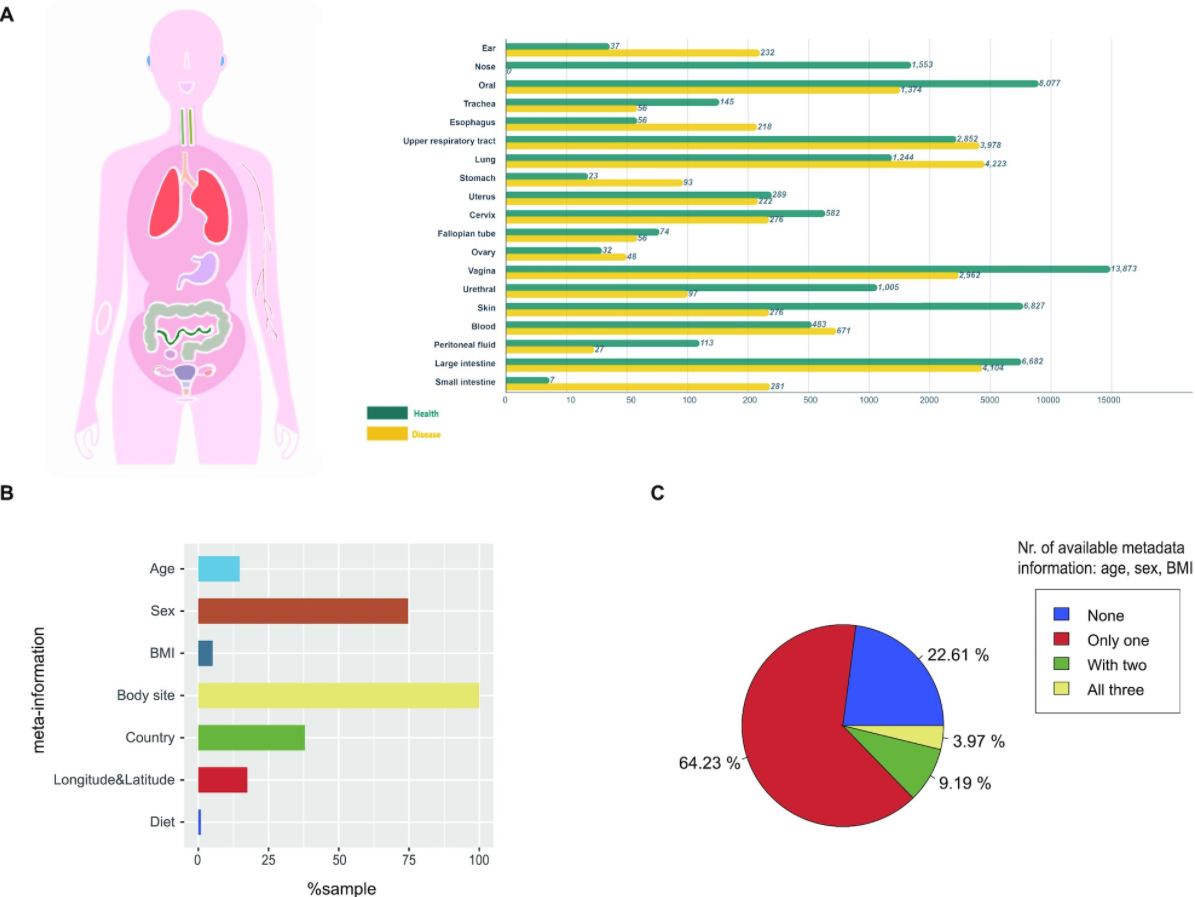
13. mBodyMap: a curated database for microbes across human body and their associations with health and diseases
by Hanbo Jin, Guoru Hu, Chuqing Sun, Yiqian Duan, Zhenmo Zhang, Zhi Liu, Xing-Ming Zhao, Wei-Hua Chen in Nucleic Acids Research
DOI: 10.1093/nar/gkab973
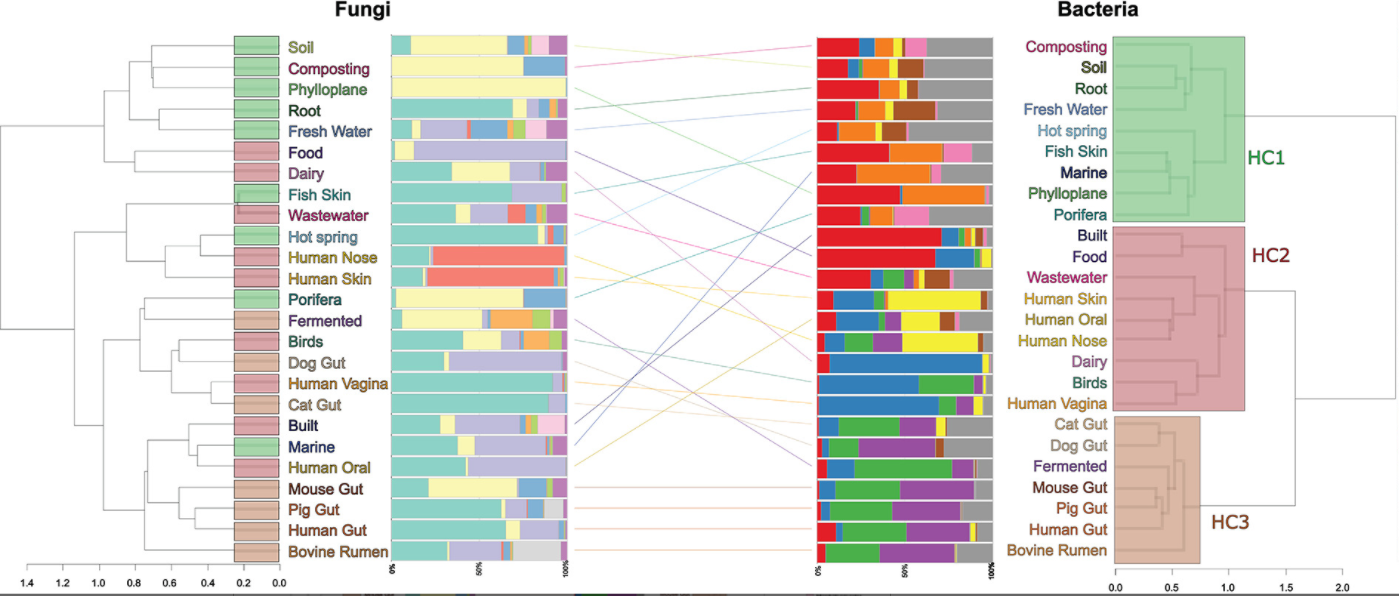
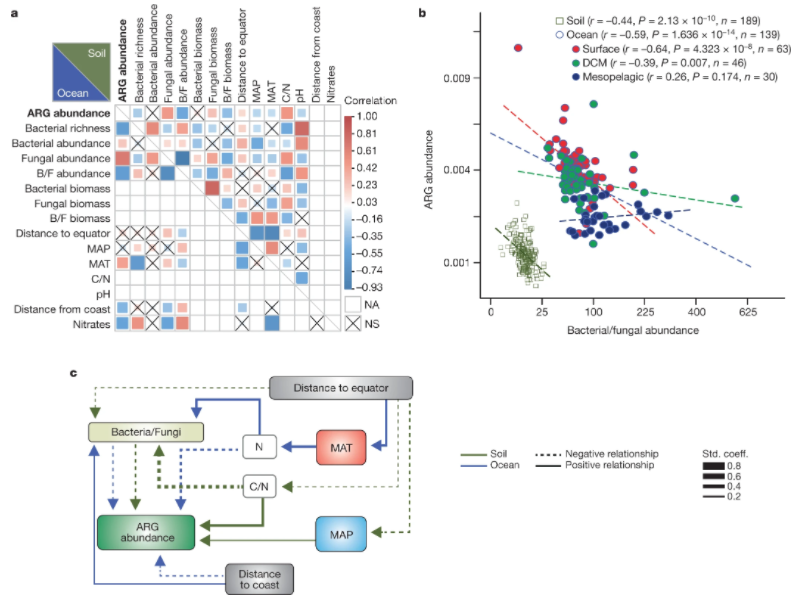
12. Metagenomic assessment of the global diversity and distribution of bacteria and fungi
by Mohammad Bahram, Tarquin Netherway, Clemence Frioux, Pamela Ferretti, Luis Pedro Coelho, Stefan Geisen, Peer Bork, Falk Hildebrand in Environmental Microbiology
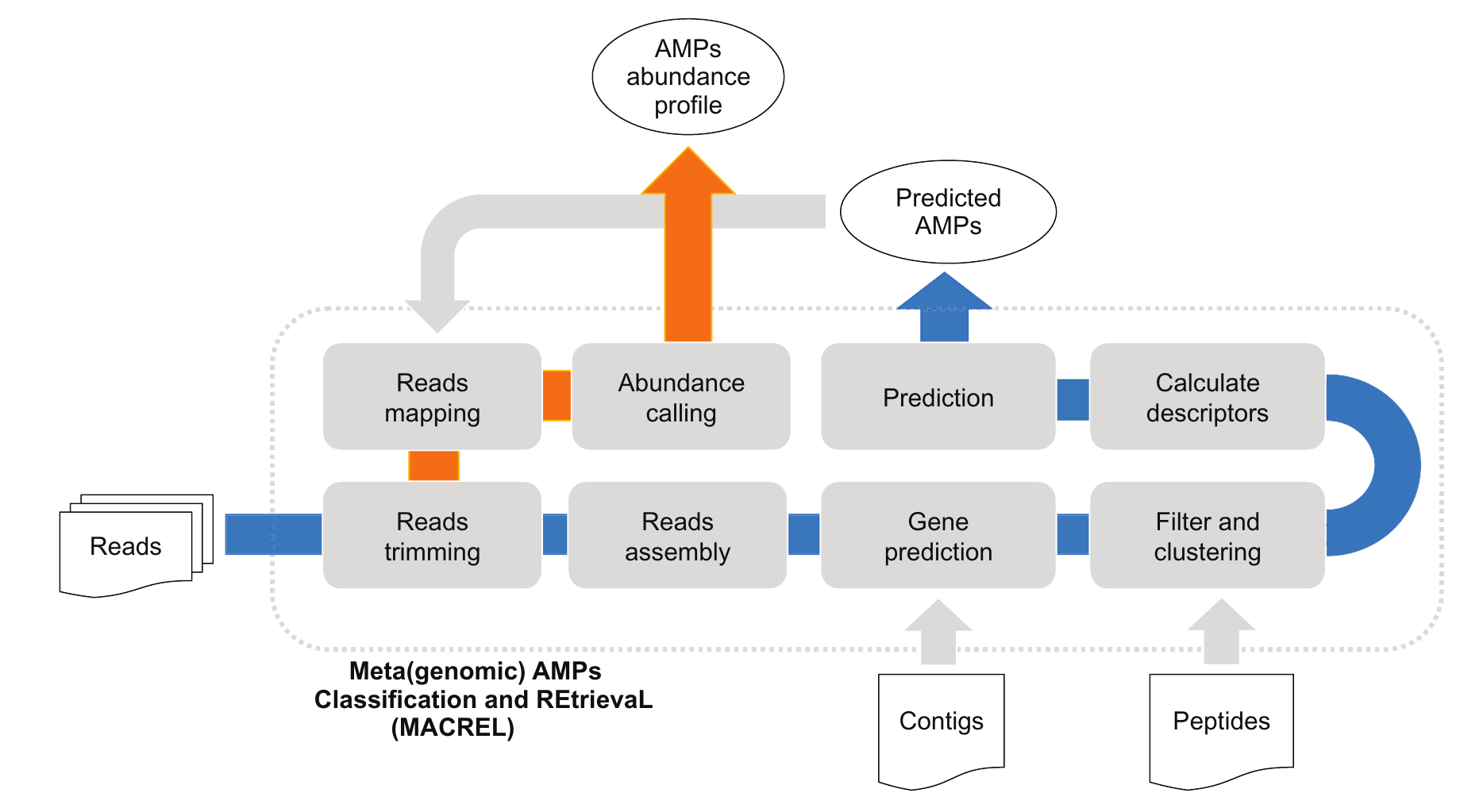
11. Macrel: antimicrobial peptide screening in genomes and metagenomes
10. Disentangling the impact of environmental and phylogenetic constraints on prokaryotic within-species diversity
by Oleksandr M. Maistrenko, Daniel R. Mende, Mechthild Luetge, ..., Luis Pedro Coelho, ..., Shinichi Sunagawa, Peer Bork in The ISME Journal
9. NG-meta-profiler: fast processing of metagenomes using NGLess, a domain-specific language
by Luis Pedro Coelho, Renato Alves, Paulo Monteiro, Jaime Huerta-Cepas, Ana Teresa Freitas, Peer Bork in Microbiome
8. Microbial abundance, activity and population genomic profiling with mOTUs2
by Alessio Milanese, Daniel R Mende, Lucas Paoli, ..., Luis Pedro Coelho, ..., Georg Zeller, Shinichi Sunagawa in Nature Communications
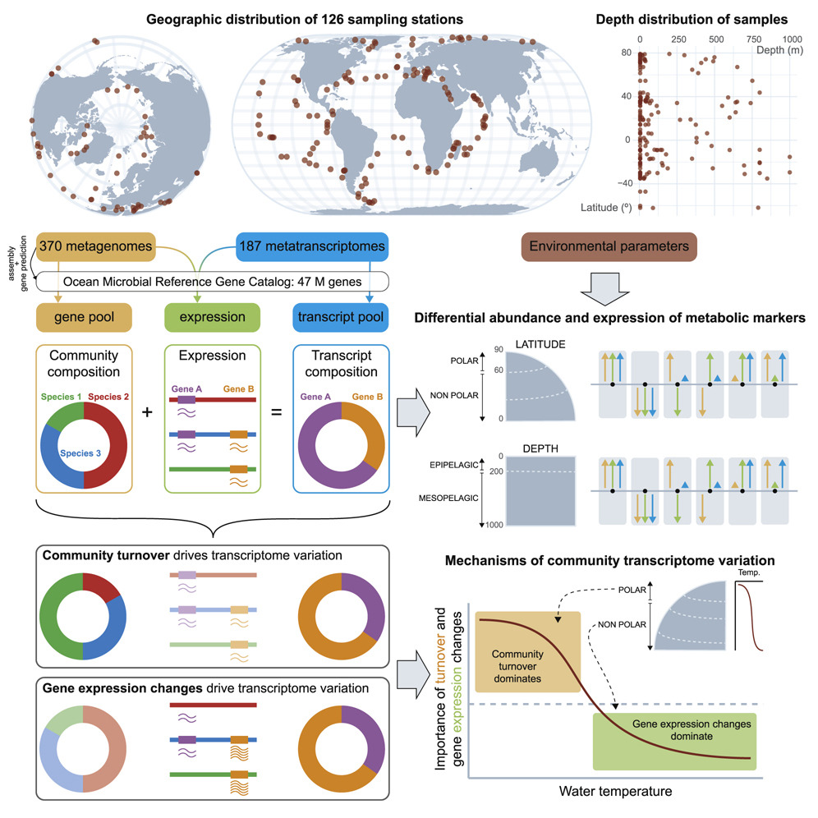
7. Gene Expression Changes and Community Turnover Differentially Shape the Global Ocean Metatranscriptome
by Guillem Salazar, Lucas Paoli, Adriana Alberti, ..., Luis Pedro Coelho, ..., Shinichi Sunagawa, Patrick Wincker in Cell
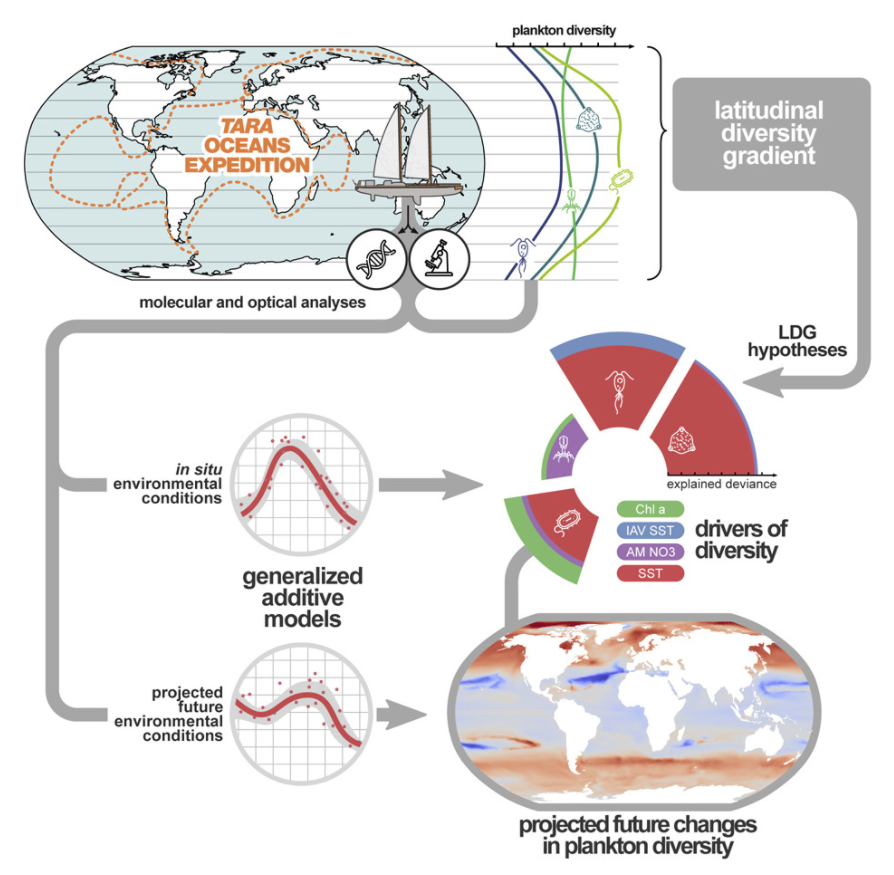
6. Global Trends in Marine Plankton Diversity across Kingdoms of Life
by Federico M. Ibarbalz, Nicolas Henry, Manoela C. Brandao, ..., Luis Pedro Coelho, ..., Shinichi Sunagawa, Patrick Wincker in Cell
5. proGenomes2: an improved database for accurate and consistent habitat, taxonomic and functional annotations of prokaryotic genomes
by Daniel R Mende, Ivica Letunic, Oleksandr M Maistrenko, ..., Luis Pedro Coelho, Peer Bork in Nucleic Acids Research
DOI: 10.1093/nar/gkz1002
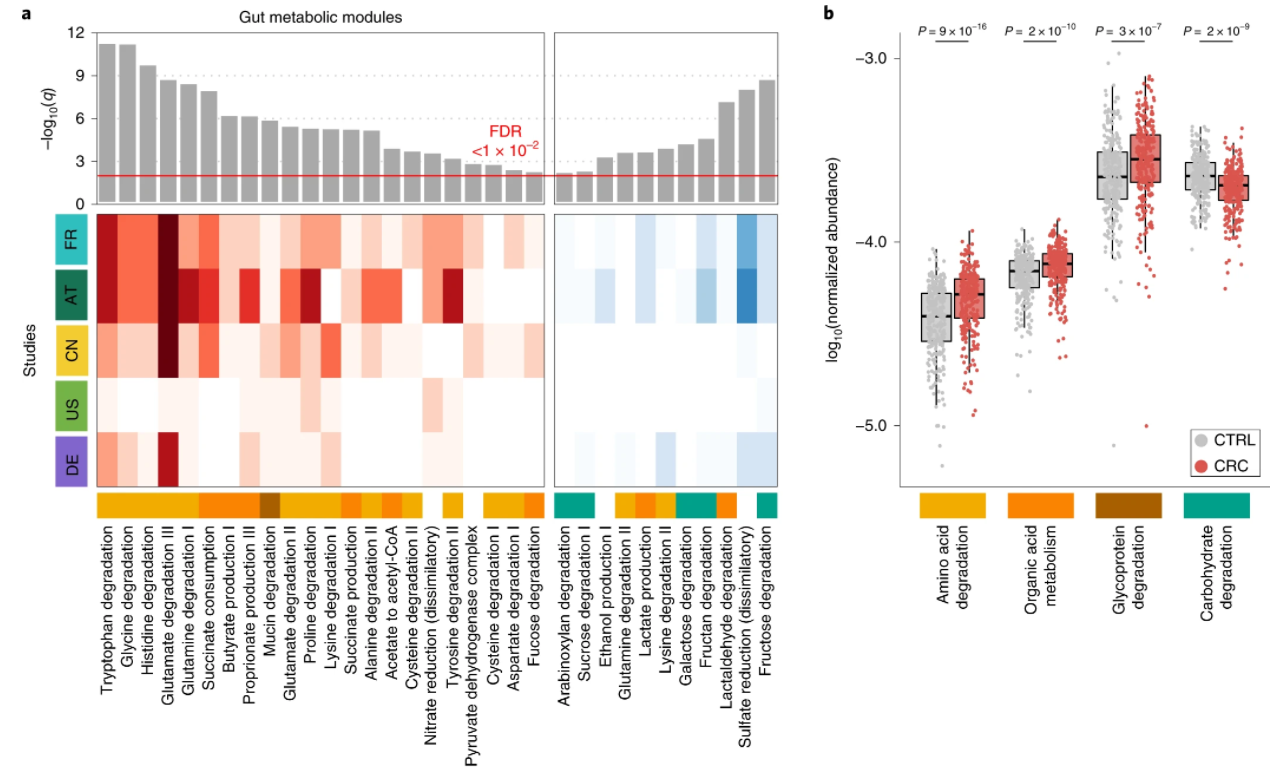
4. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer
by Jakob Wirbel, Paul Theodor Pyl, Ece Kartal, ..., Luis Pedro Coelho, ..., Peer Bork, Georg Zeller in Nature Medicine
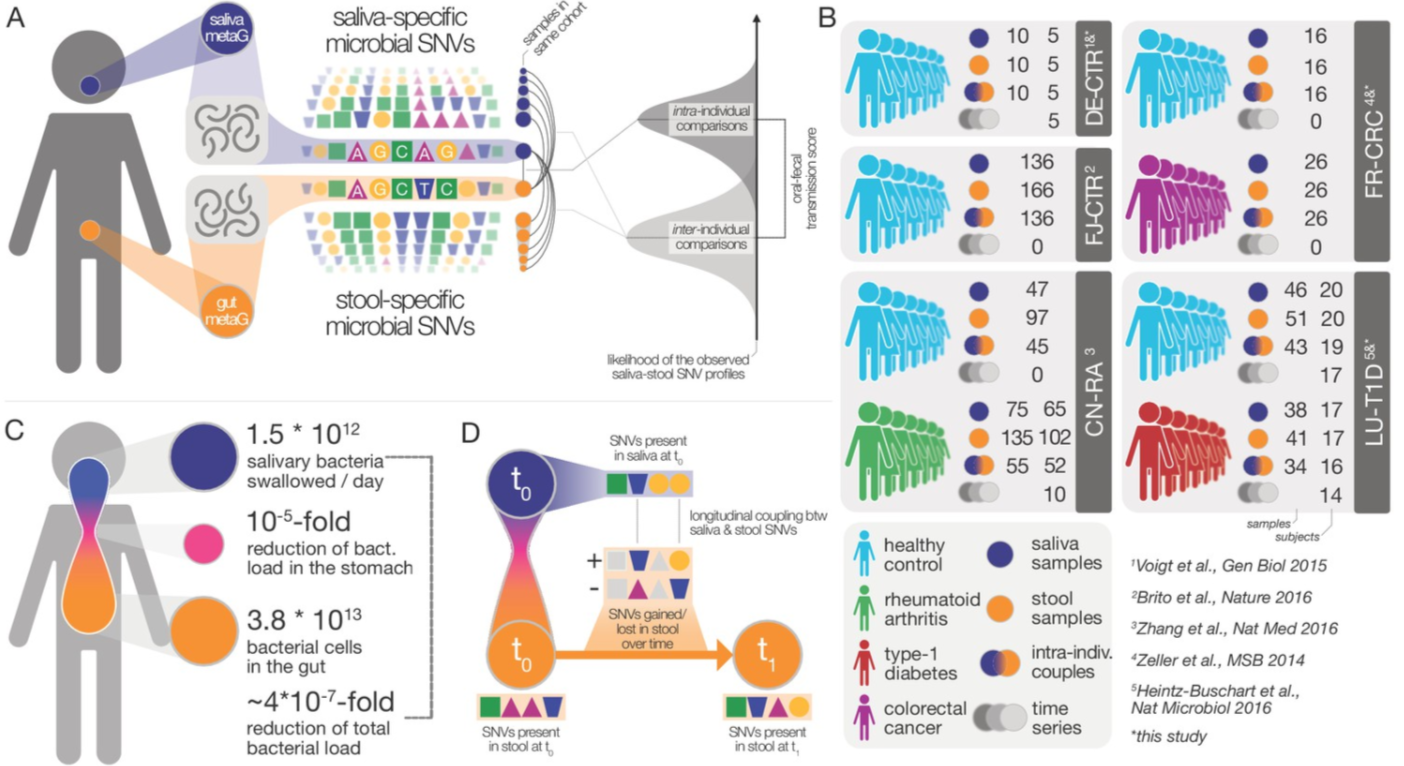
3. Extensive transmission of microbes along the gastrointestinal tract
by Thomas SB Schmidt, Matthew R Hayward, Luis Pedro Coelho, ..., Paul Wilmes, Peer Bork in eLife
DOI: 10.7554/elife.42693
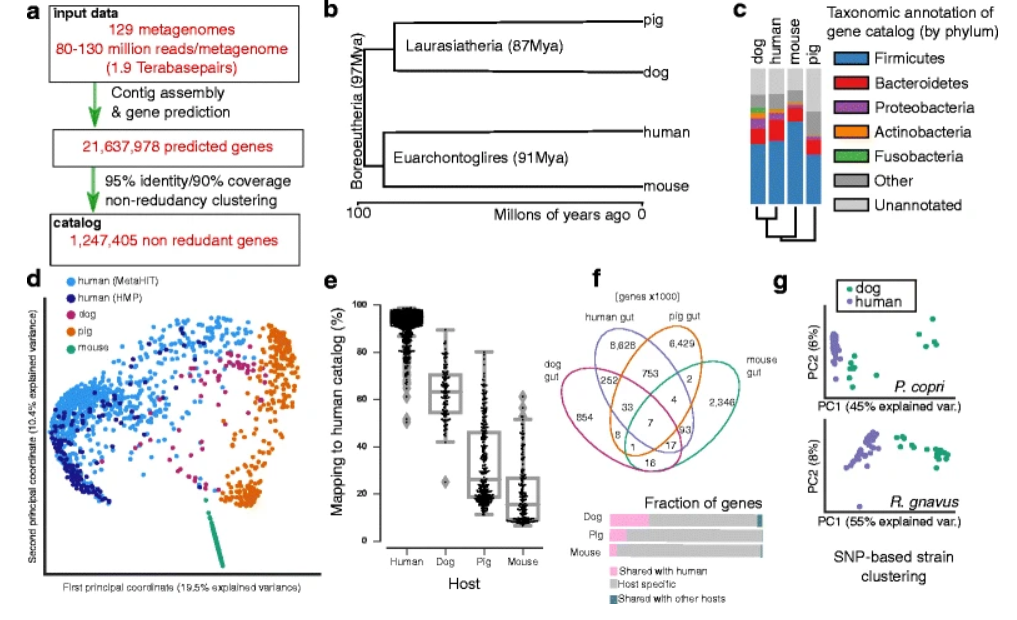
2. Similarity of the dog and human gut microbiomes in gene content and response to diet
by Luis Pedro Coelho, Jens Roat Kultima, Paul Igor Costea, ..., Qinghong Li, Peer Bork in Microbiome
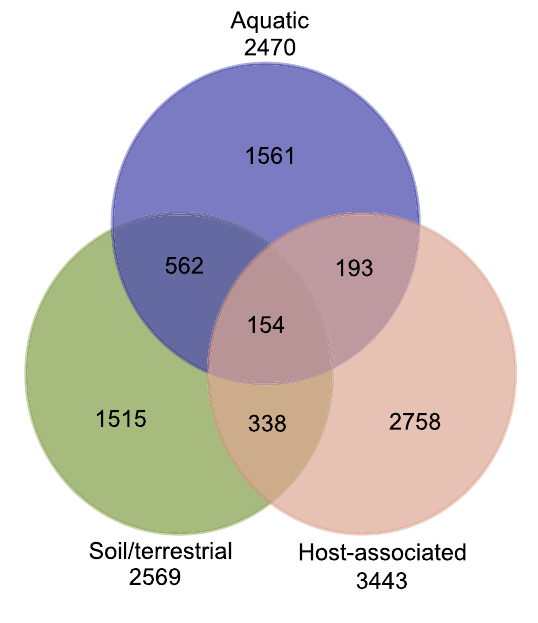
1. Structure and function of the global topsoil microbiome
by Mohammad Bahram, Falk Hildebrand, Sofia K. Forslund, ..., Luis Pedro Coelho, ..., Leho Tedersoo, Peer Bork in Nature
Copyright (c) 2018–2024. Luis Pedro Coelho and other group members. All rights reserved.