BDB-Lab members involved
metaMIC: reference-free Misassembly Identification and Correction of de novo metagenomic assemblies
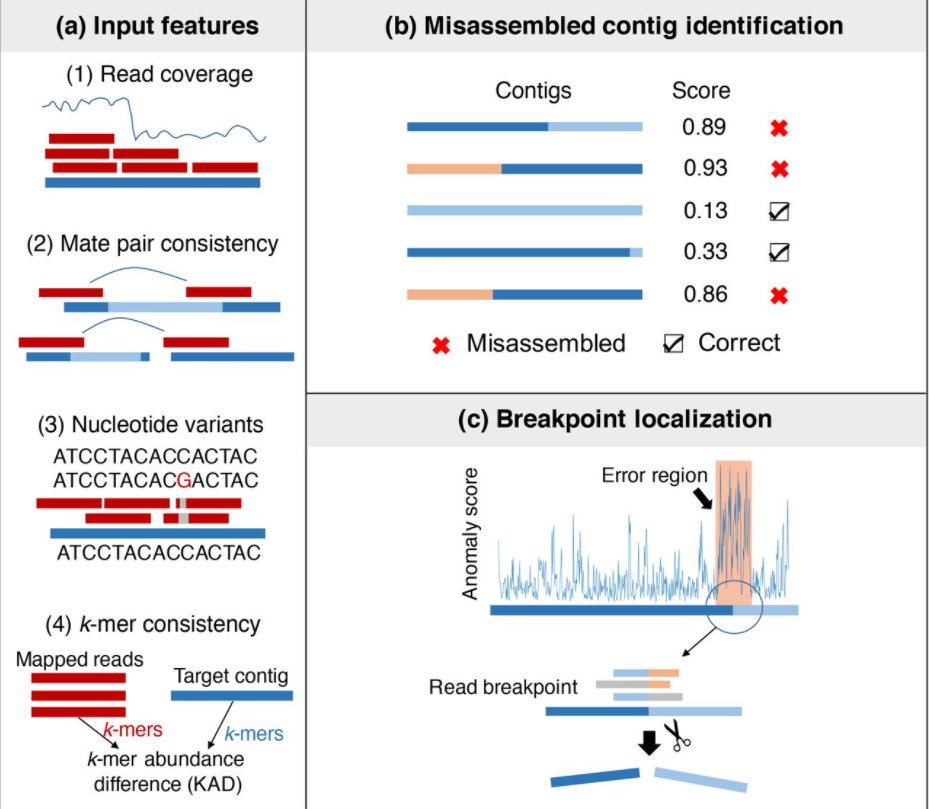
metaMIC: reference-free Misassembly Identification and Correction of de novo metagenomic assemblies
by Senying Lai, Shaojun Pan, Chuqing Sun, Luis Pedro Coelho, Wei-Hua Chen, Xing-Ming Zhao
Abstract
Evaluating the quality of metagenomic assemblies is important for constructing reliable metagenome-assembled genomes and downstream analyses. Here, we present metaMIC (https://github.com/ZhaoXM-Lab/metaMIC), a machine-learning based tool for identifying and correcting misassemblies in metagenomic assemblies. Benchmarking results on both simulated and real datasets demonstrate that metaMIC outperforms existing tools when identifying misassembled contigs. Furthermore, metaMIC is able to localize the misassembly breakpoints, and the correction of misassemblies by splitting at misassembly breakpoints can improve downstream scaffolding and binning results.
Full text: https://doi.org/10.1101/2021.06.22.449514
Copyright (c) 2018–2024. Luis Pedro Coelho and other group members. All rights reserved.
