Most recent BDB-Lab papers
BDB-Lab Links
Major Projects
Other Links
Lab Members
Twitter feed
Tweets by BigDataBiologyPublications
41. Persistence of High-Risk Antimicrobial Resistance Genes in Extracellular DNA Along an Urban Wastewater-River Continuum
by John Paul Makumbi, Samuel K. Leareng, Oliver K. Bezuidt, Luis Pedro Coelho, Thulani P. Makhalanyane in Sneak Peek (SSRN Preprints)
This study investigates the persistence of high-risk antimicrobial resistance (AMR) genes in extracellular DNA along an urban wastewater-river continuum, highlighting the potential for environmental transmission of AMR.
DOI: 10.2139/ssrn.5342551
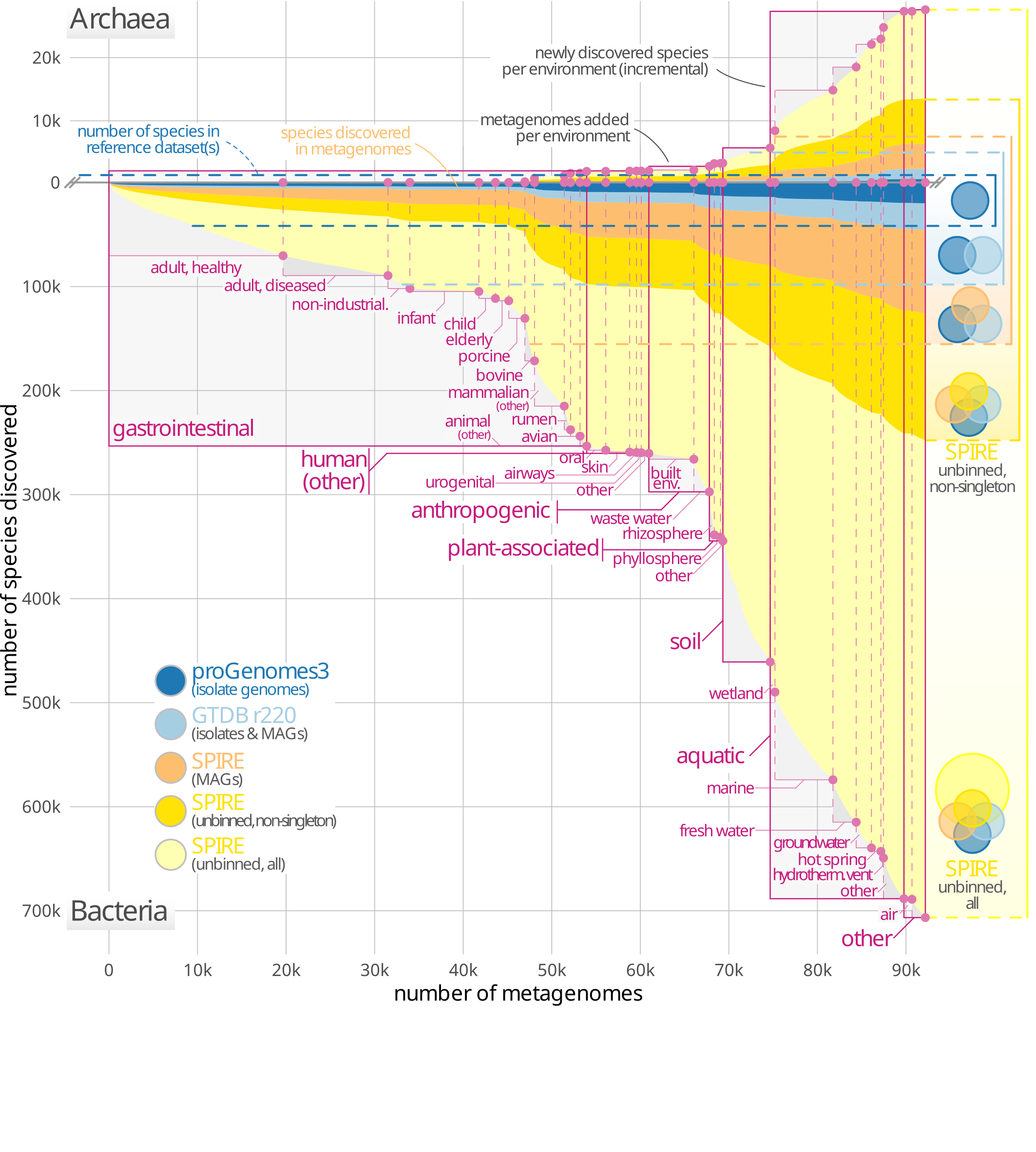
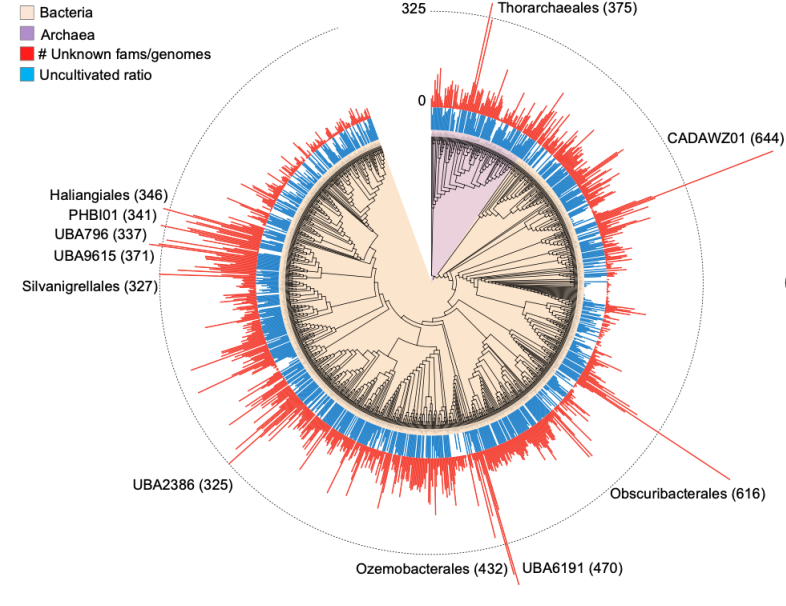
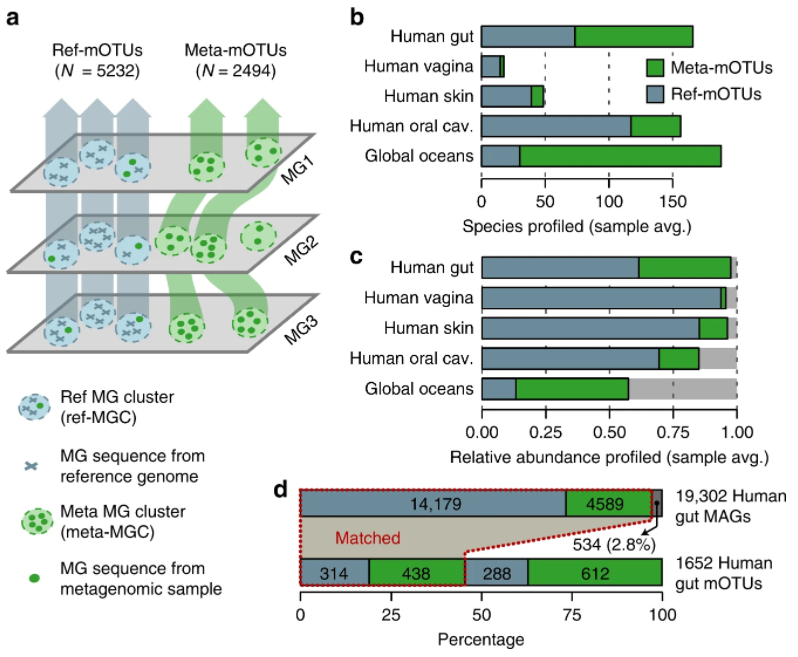
40. A census of hidden and discoverable microbial diversity beyond genome-centric approaches
by Vishnu Prasoodanan P K, Oleksandr M Maistrenko, Anthony Fullam, Daniel R Mende, Ece Kartal, Luis Pedro Coelho, Anja Spang, Peer Bork, Thomas S B Schmidt in bioRxiv (PREPRINT)
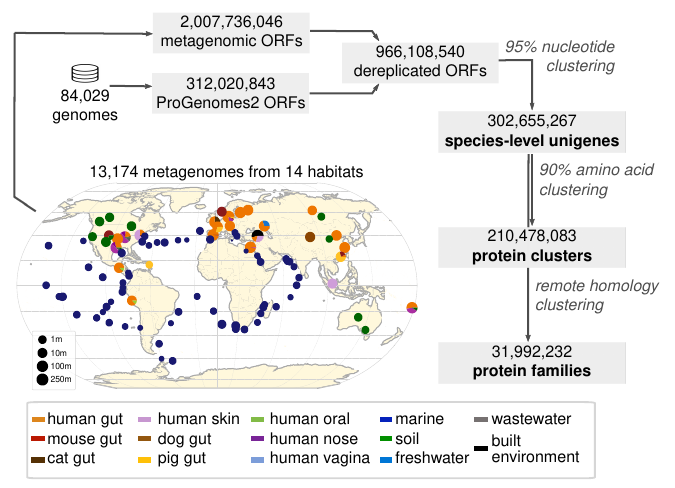
We investigated the discoverable bacterial and archaeal diversity in a cross-habitat dataset of 92,187 metagenomes, revealing a vast majority of species-level clades hidden among unbinned contigs. We predict that discovery of novel microbial lineages will continue across habitats as more data accrues for the foreseeable future.
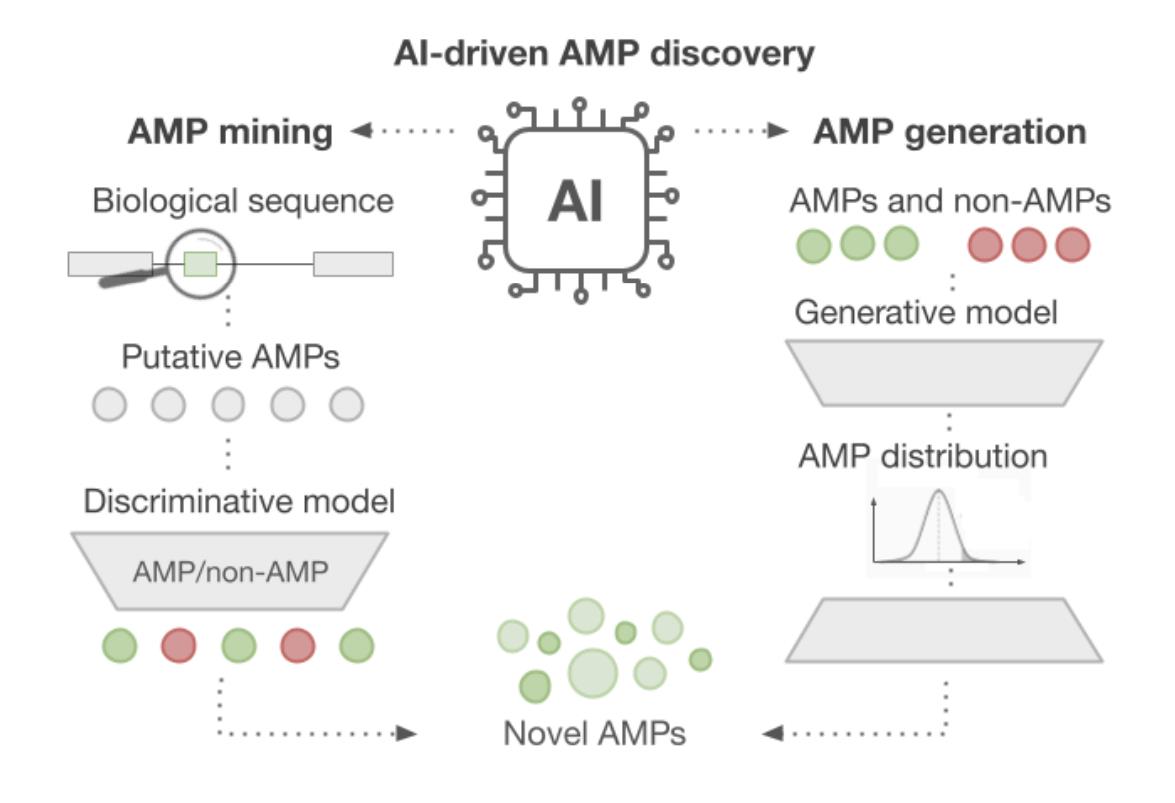
39. AI-Driven Antimicrobial Peptide Discovery: Mining and Generation
by Paulina Szymczak, Wojciech Zarzecki, Jiejing Wang, Yiqian Duan, Jun Wang, Luis Pedro Coelho, Cesar de la Fuente-Nunez, Ewa Szczurek in Accounts of Chemical Research
Describe the technical challenges and advancements with AI-based approaches in antimicrobial peptide discovery.
38. argNorm: Normalization of antibiotic resistance gene annotations to the Antibiotic Resistance Ontology (ARO)
by Svetlana Ugarcina Perovic, Vedanth Ramji, Hui Chong, Yiqian Duan, Finlay Maguire, Luis Pedro Coelho in Bioinformatics
Presenting argNorm, a command line tool and Python library to normalize antibiotic resistance gene annotations to the Antibiotic Resistance Ontology (ARO)
37. Quest for Orthologs in the Era of Biodiversity Genomics
by Felix Langschied, Nicola Bordin, Salvatore Cosentino, ..., Luis Pedro Coelho, ..., Christophe Dessimoz, Ingo Ebersberger in Genome Biology and Evolution
We discuss challenges and opportunities for orthology inference in the context of the ongoing biodiversity genomics revolution.
DOI: 10.1093/gbe/evae224
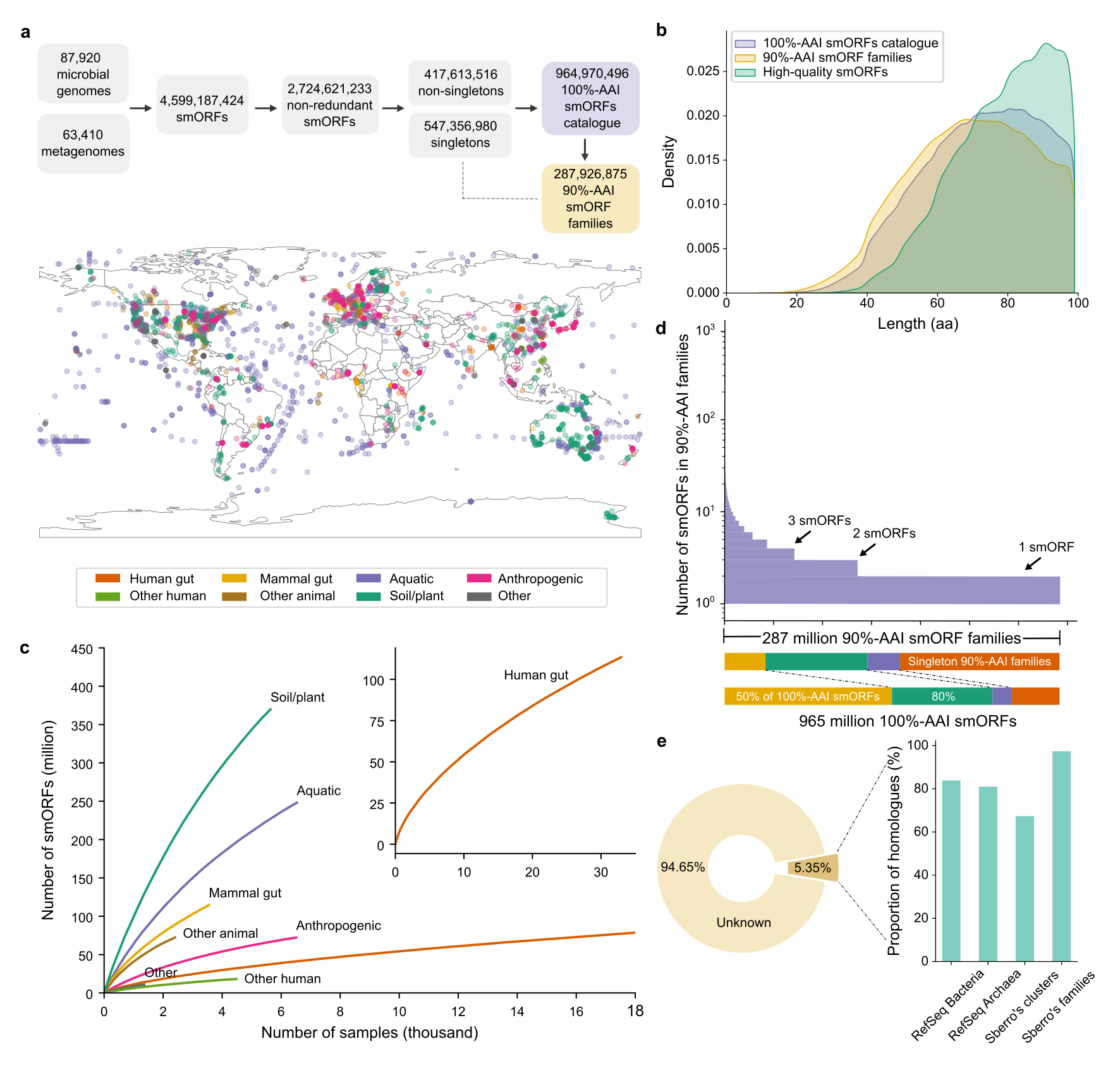
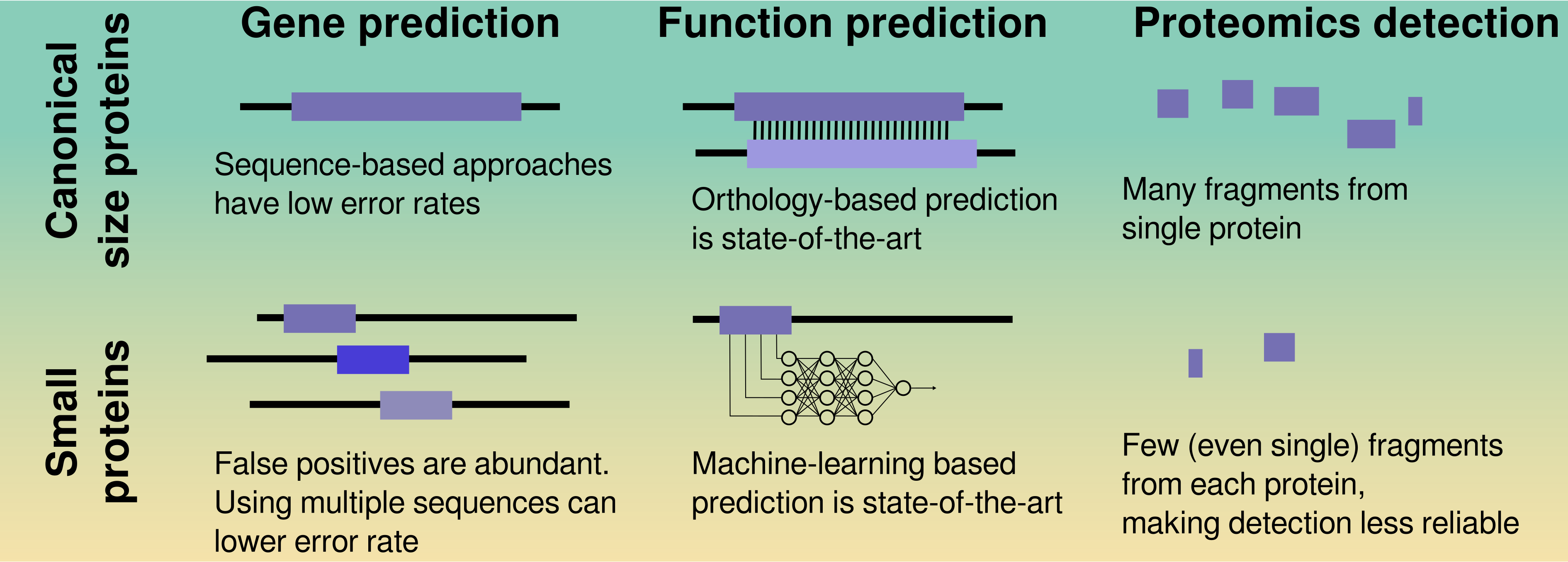
36. A catalog of small proteins from the global microbiome
by Yiqian Duan, Célio Dias Santos Júnior, Thomas S.B. Schmidt, Anthony Fullam, Breno L. S. de Almeida, Chengkai Zhu, Kuhn Michael, Xing-Ming Zhao, Peer Bork, Luis Pedro Coelho in Nature Communications
We constructed a global microbial smORFs catalog (GMSC), demonstrating the immense and underexplored diversity of small proteins in the microbiome.
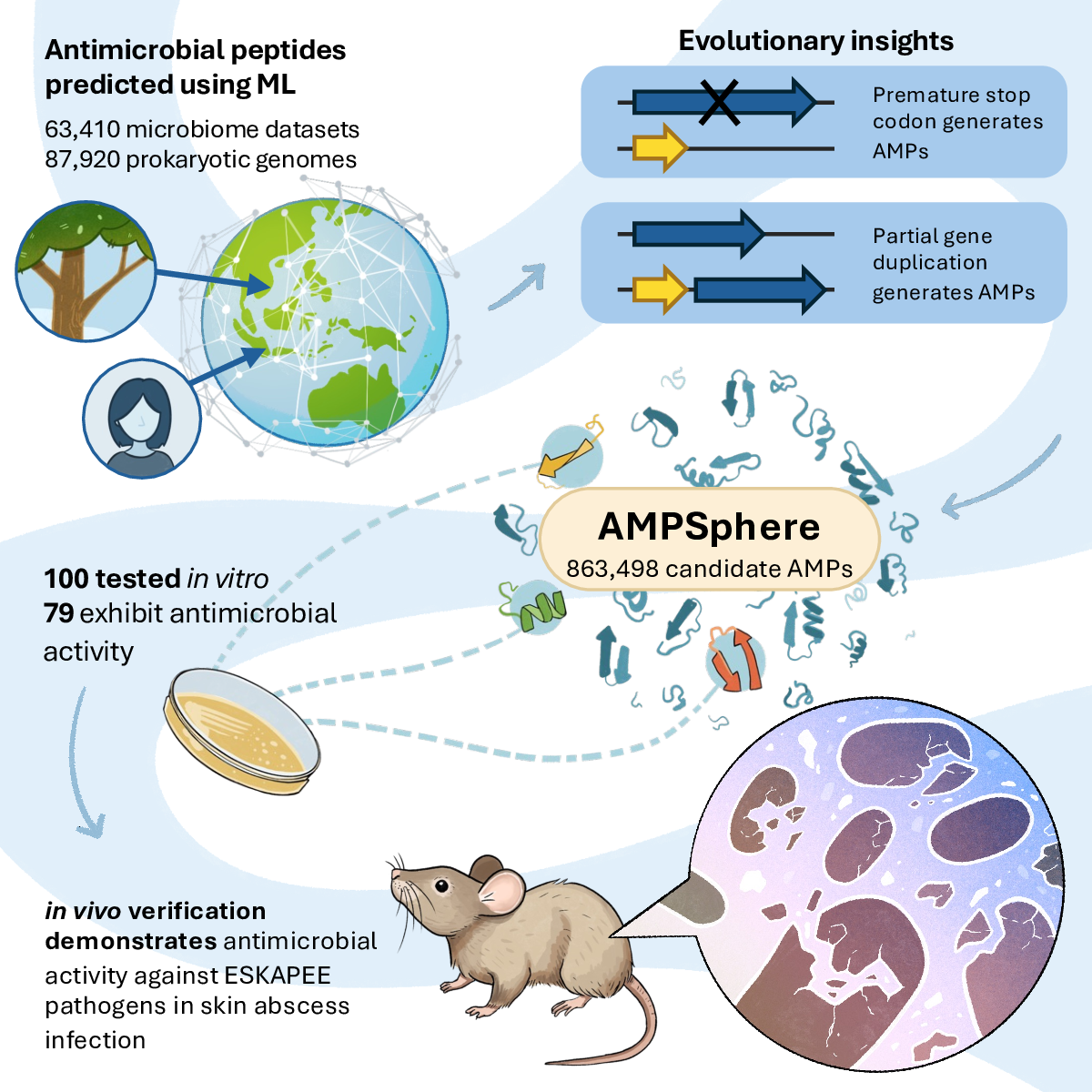
35. Discovery of antimicrobial peptides in the global microbiome with machine learning
by Célio Dias Santos Júnior, Marcelo D.T. Torres, Yiqian Duan, ..., Hui Chong, ..., Chengkai Zhu, Amy Houseman, ..., Cesar de la Fuente-Nunez, Luis Pedro Coelho in Cell
AMPSphere is an exploration of the antimicrobial peptides (AMPs) of the global microbiome.
34. For long-term sustainable software in bioinformatics
by Luis Pedro Coelho in PLOS Computational Biology
We propose a framework for categorizing research software by its intended use and maintenance level and share seven practices for ensuring long-term maintenance, including reproducible research methods and public support forums. We call for the bioinformatics community to institutionalize software maintenance commitments through funders, peer reviewers, and journals, similar to data sharing mandates, to ensure software's long-term viability and reproducibility.
33. Challenges in computational discovery of bioactive peptides in ’omics data
by Luis Pedro Coelho, Célio Dias Santos Júnior, Cesar de la Fuente‐Nunez in PROTEOMICS
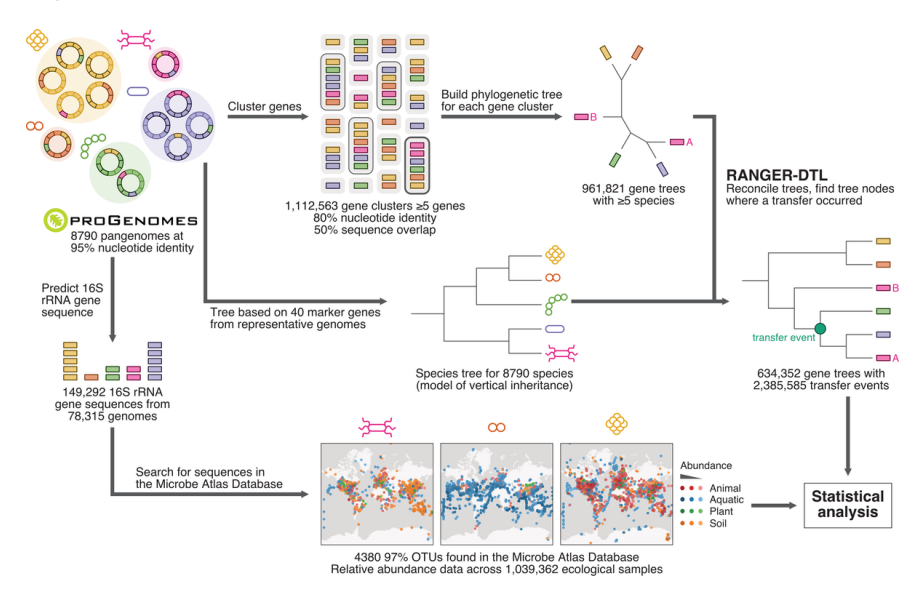
32. A global survey of prokaryotic genomes reveals the eco-evolutionary pressures driving horizontal gene transfer
by Marija Dmitrijeva, Janko Tackmann, João Frederico Matias Rodrigues, Jaime Huerta-Cepas, Luis Pedro Coelho, Christian von Mering in Nature Ecology & Evolution
We studied horizontal gene transfer (HGT) using tree reconcilation between gene and species trees, which enabled us to observe different profiles for recent versus old events. For example, genes transferred earlier in evolution tend to be more ubiquitous within present-day species. We also found that co-occurring, interacting, and high-abundance species tend to exchange more genes.
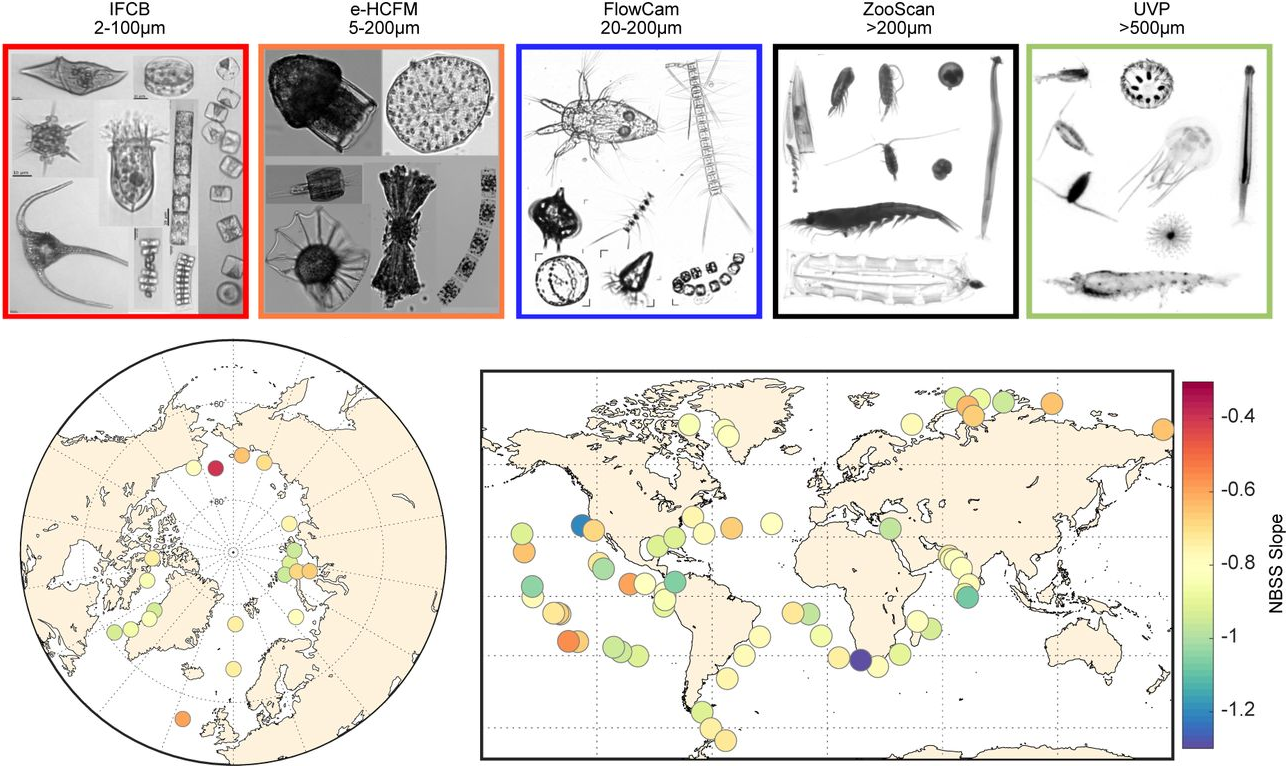
31. Ubiquity of inverted ’gelatinous’ ecosystem pyramids in the global ocean
by Fabien Lombard, Lionel Guidi, Manoela C. Brandão, Luis Pedro Coelho, ..., Gabriel Gorsky, Tara Oceans Coordinators in bioRxiv (PREPRINT)
Using a multitude of imaging methods, we cover 5 orders of magnitude to reveal that gelatinous organisms represent 30% of the total marine plantkon biovolume.
30. Functional and evolutionary significance of unknown genes from uncultivated taxa
by Álvaro Rodríguez del Río, Joaquín Giner-Lamia, Carlos P. Cantalapiedra, ..., Luis Pedro Coelho, Jaime Huerta-Cepas in Nature
Thousands of previously-ignored protein families are found across the global microbiome.
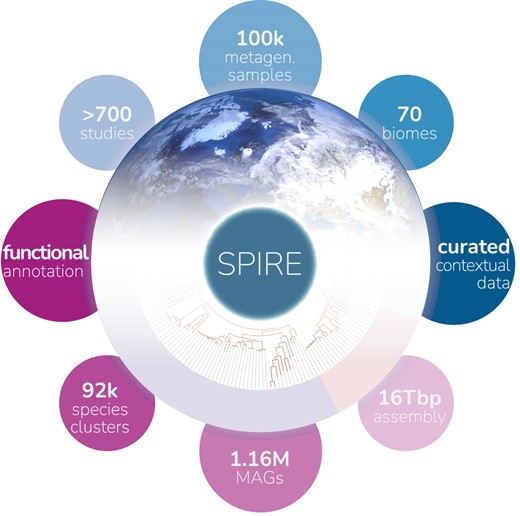
29. SPIRE: a Searchable, Planetary-scale mIcrobiome REsource
by Thomas S B Schmidt, Anthony Fullam, Pamela Ferretti, ..., Yiqian Duan, ..., Luis Pedro Coelho, Peer Bork in Nucleic Acids Research
SPIRE is a Searchable Planetary-scale mIcrobiome REsource that integrates various consistently processed metagenome-derived microbial data modalities across habitats, geography and phylogeny.
DOI: 10.1093/nar/gkad943
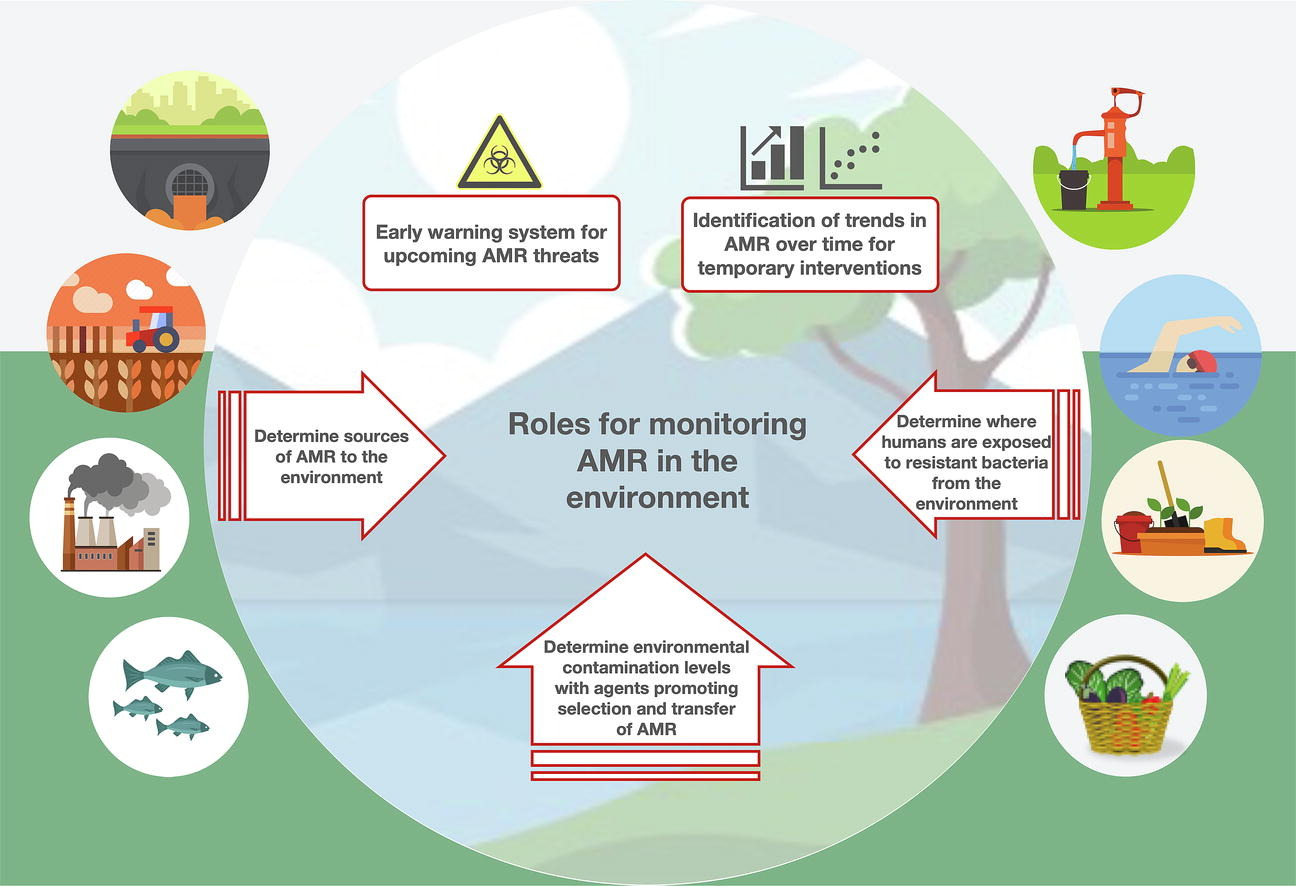
28. Towards Monitoring of Antimicrobial Resistance in the Environment: For what Reasons, How to Implement It, and What Are the Data Needs?
by Johan Bengtsson-Palme, Anna Abramova, Thomas U. Berendonk, Luis Pedro Coelho, ..., Svetlana Ugarcina Perovic, ..., Astrid Louise Wester, Rabaab Zahra in Environment International
Environmental monitoring of antimicrobial resistance could help fight the resistance crisis, but some challenges remain
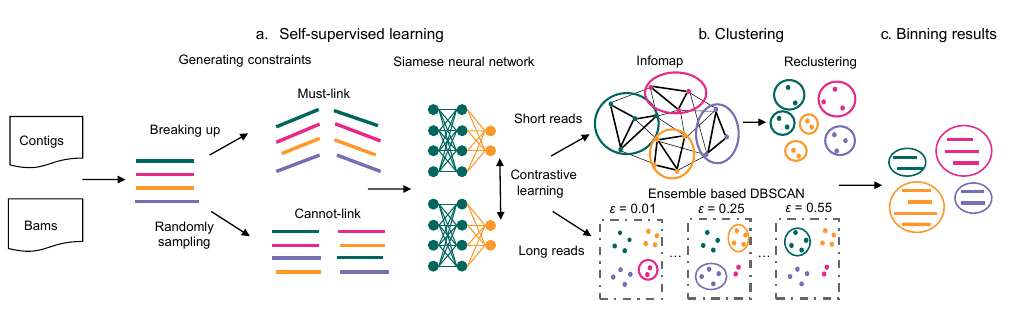
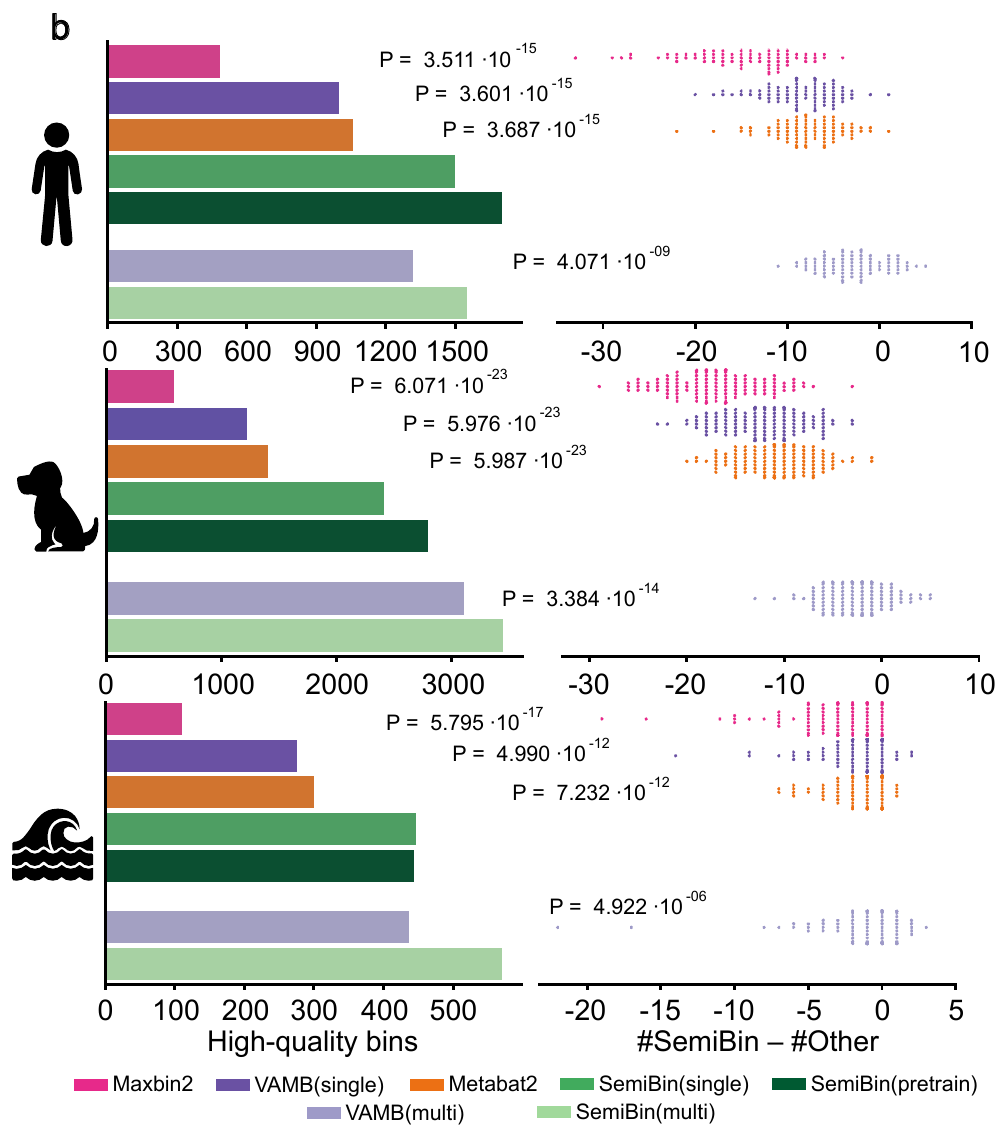
27. SemiBin2: self-supervised contrastive learning leads to better MAGs for short- and long-read sequencing
by Shaojun Pan, Xing-Ming Zhao, Luis Pedro Coelho in Bioinformatics
SemiBin2 uses self-supervised learning to improve on SemiBin1. It additionally adds support for long-read sequencing data.
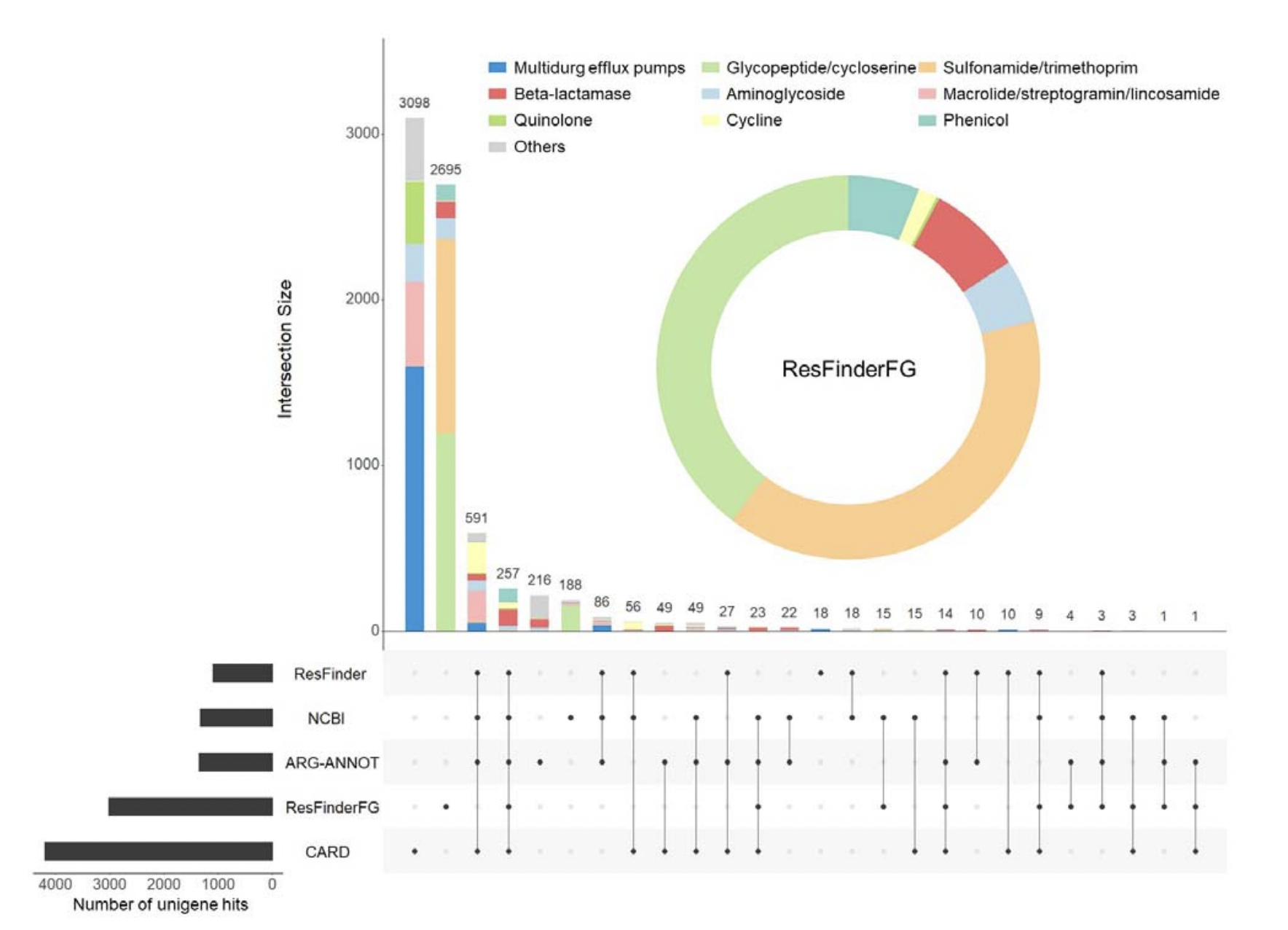
26. ResFinderFG v2.0: a database of antibiotic resistance genes obtained by functional metagenomics
by Rémi Gschwind, Svetlana Ugarcina Perovic, Maja Weiss, Marie Petitjean, Julie Lao, Luis Pedro Coelho, Etienne Ruppé in Nucleic Acids Research
ResFinderFG version 2
DOI: 10.1093/nar/gkad384
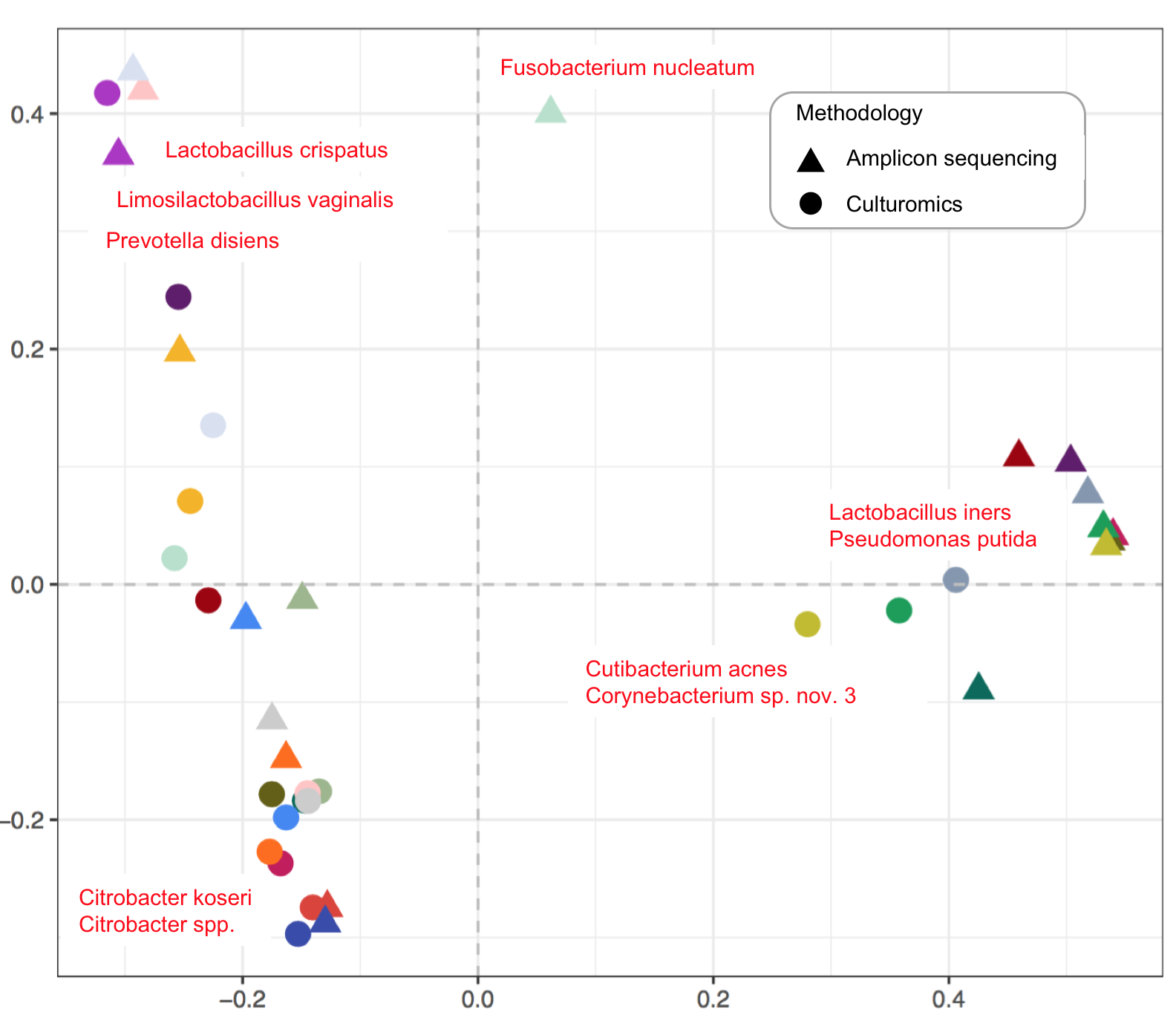
25. Urinary Microbiome of Reproductive-Age Asymptomatic European Women
by Svetlana Ugarcina Perovic, Magdalena Ksiezarek, Joana Rocha, Elisabete Alves Cappelli, Márcia Sousa, Teresa Gonçalves Ribeiro, Filipa Grosso, Luísa Peixe in Microbiology Spectrum
Baterial diversity in healthy female urinary tract
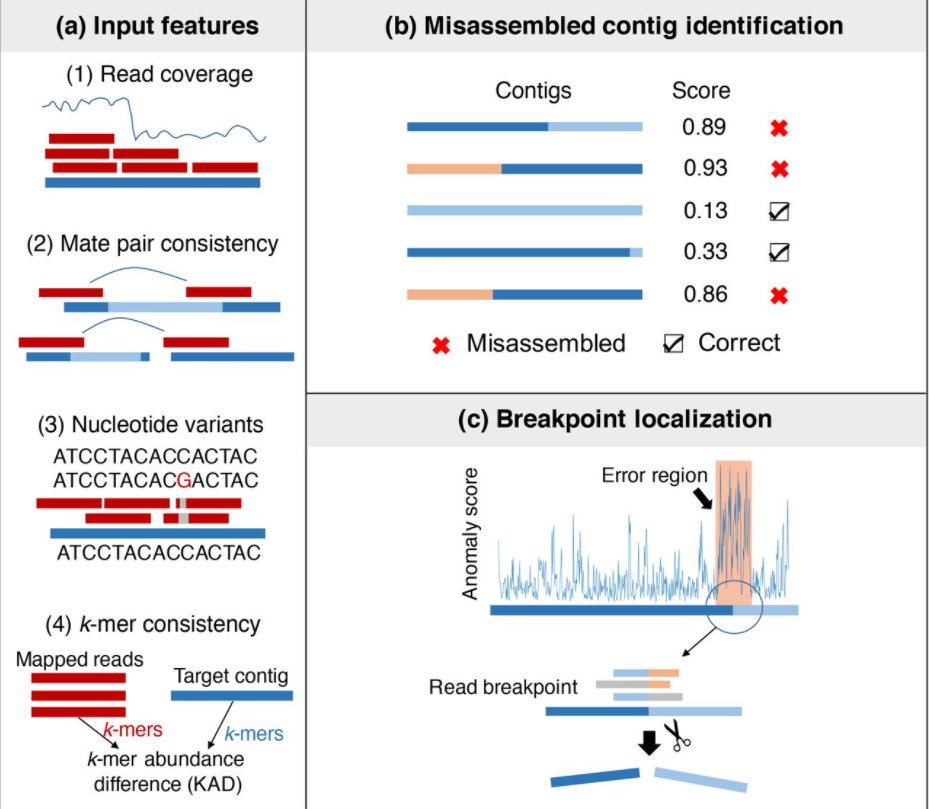
24. metaMIC: reference-free Misassembly Identification and Correction of de novo metagenomic assemblies
by Senying Lai, Shaojun Pan, Chuqing Sun, Luis Pedro Coelho, Wei-Hua Chen, Xing-Ming Zhao in Genome biology
A machine-learning based tool for identifying and correcting misassemblies in metagenomic assemblies.
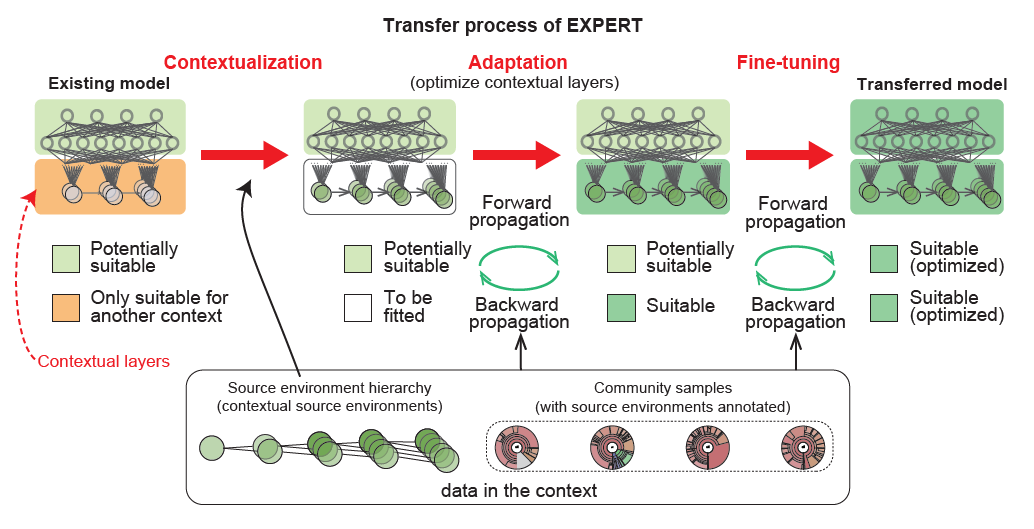
23. EXPERT: Transfer Learning-enabled context-aware microbial source tracking
by Hui Chong, Yuguo Zha, Qingyang Yu, ..., Luis Pedro Coelho, Kang Ning in Briefings in Bioinformatics
Transfer Learning-enabled context-aware microbial source tracking
DOI: 10.1093/bib/bbac396
22. Drivers and Determinants of Strain Dynamics Following Faecal Microbiota Transplantation
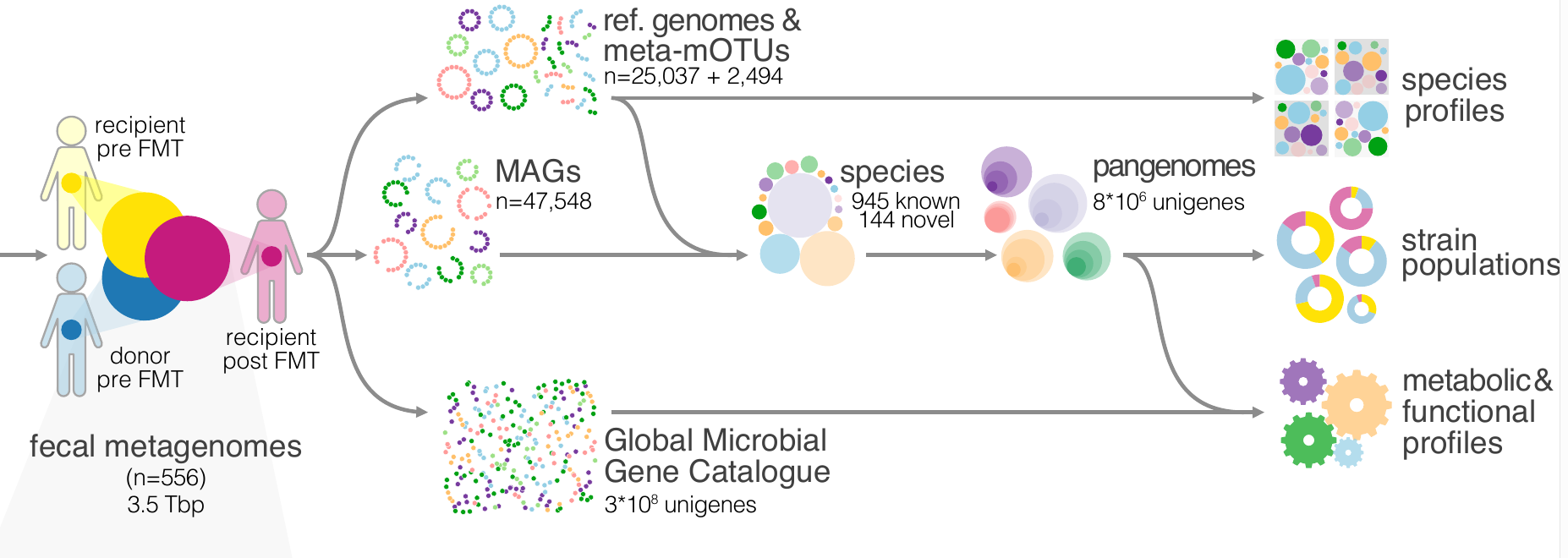
[More about Drivers and Determinants of Strain Dynamics Following Faecal Microbiota Transplantation]
by Thomas SB Schmidt, Simone S Li, Oleksandr M Maistrenko, ..., Luis Pedro Coelho, ..., Max Nieuwdorp, Peer Bork in Nature Medicine
What happens after a fecal microbiota transplantation (FMT)? Which microbes stay and which get replaced?
21. A deep siamese neural network improves metagenome-assembled genomes in microbiome datasets across different environments
by Shaojun Pan, Chengkai Zhu, Xing-Ming Zhao, Luis Pedro Coelho in Nature Communications
SemiBin is a semi-supervised binning method for metagenomes that improves on the state of the art across multiple environments.
20. Software testing in microbial bioinformatics: a call to action
by Boas C.L. van der Putten, C. I. Mendes, Brooke M. Talbot, Jolinda de Korne-Elenbaas, Rafael Mamede, Pedro Vila-Cerqueira, Luis Pedro Coelho, Christopher A. Gulvik, Lee S. Katz, The ASM NGS 2020 Hackathon participants in Microbial Genomics
Here, we report seven recommendations that help researchers implement software testing in microbial bioinformatics.
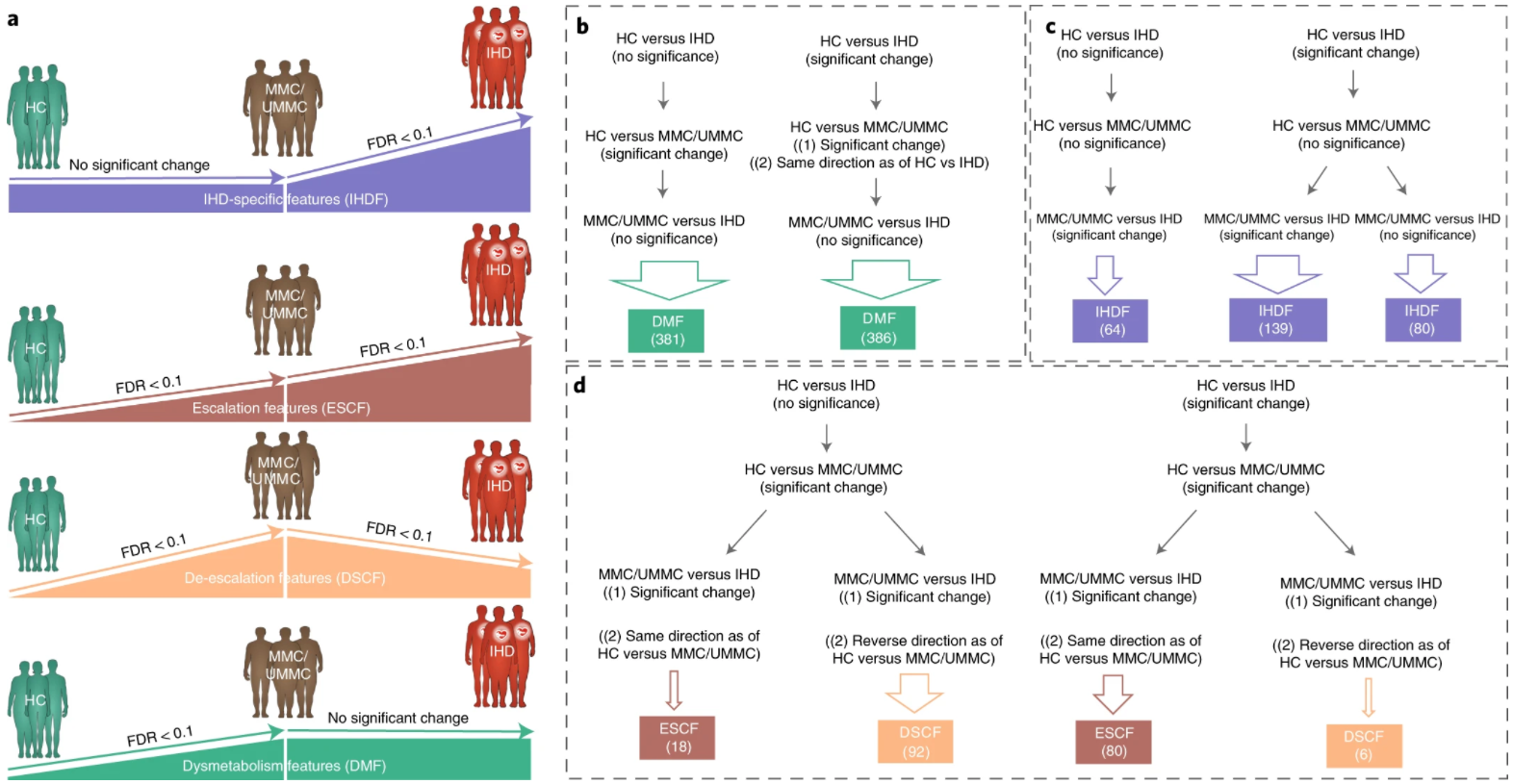
19. Microbiome and metabolome features of the cardiometabolic disease spectrum
by Sebastien Fromentin, Sofia K. Forslund, Kanta Chechi, ..., Luis Pedro Coelho, ..., S. Dusko Ehrlich, Oluf Pedersen in Nature Medicine
Segregate IHD-specific changes in gut microbial and metabolomic features from potential confounders.
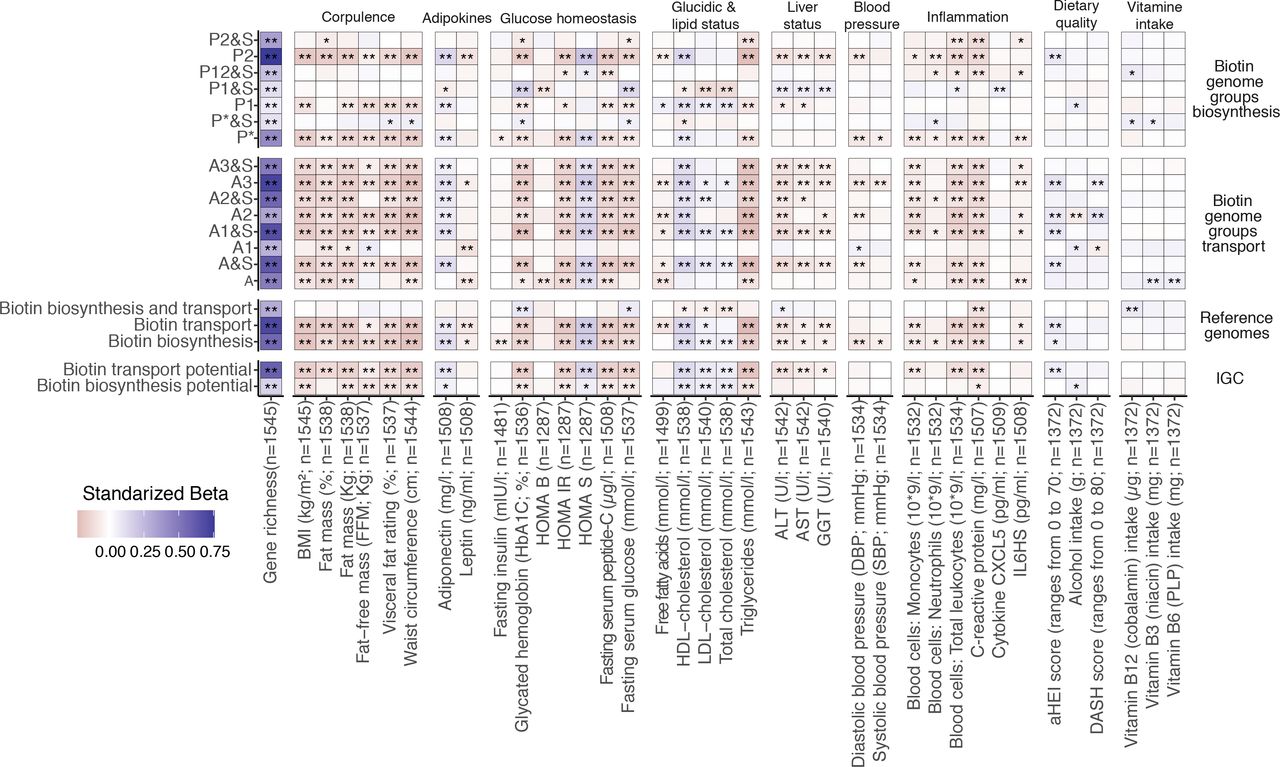
18. Impairment of gut microbial biotin metabolism and host biotin status in severe obesity: effect of biotin and prebiotic supplementation on improved metabolism
by Eugeni Belda, Lise Voland, Valentina Tremaroli, ..., Luis Pedro Coelho, ..., Karine Clément, The MetaCardis Consortium in Gut
Strategies combining biotin and prebiotic supplementation could help prevent the deterioration of metabolic states in severe obesity.
17. Towards the biogeography of prokaryotic genes
by Luis Pedro Coelho, Renato Alves, Álvaro Rodríguez del Río, ..., Shaojun Pan, ..., Jaime Huerta-Cepas, Peer Bork in Nature
Presents the GMGC and concludes that the vast genomic diversity observed is driven by neutral evolution (not adaptation).
16. Combinatorial, additive and dose-dependent drug–microbiome associations
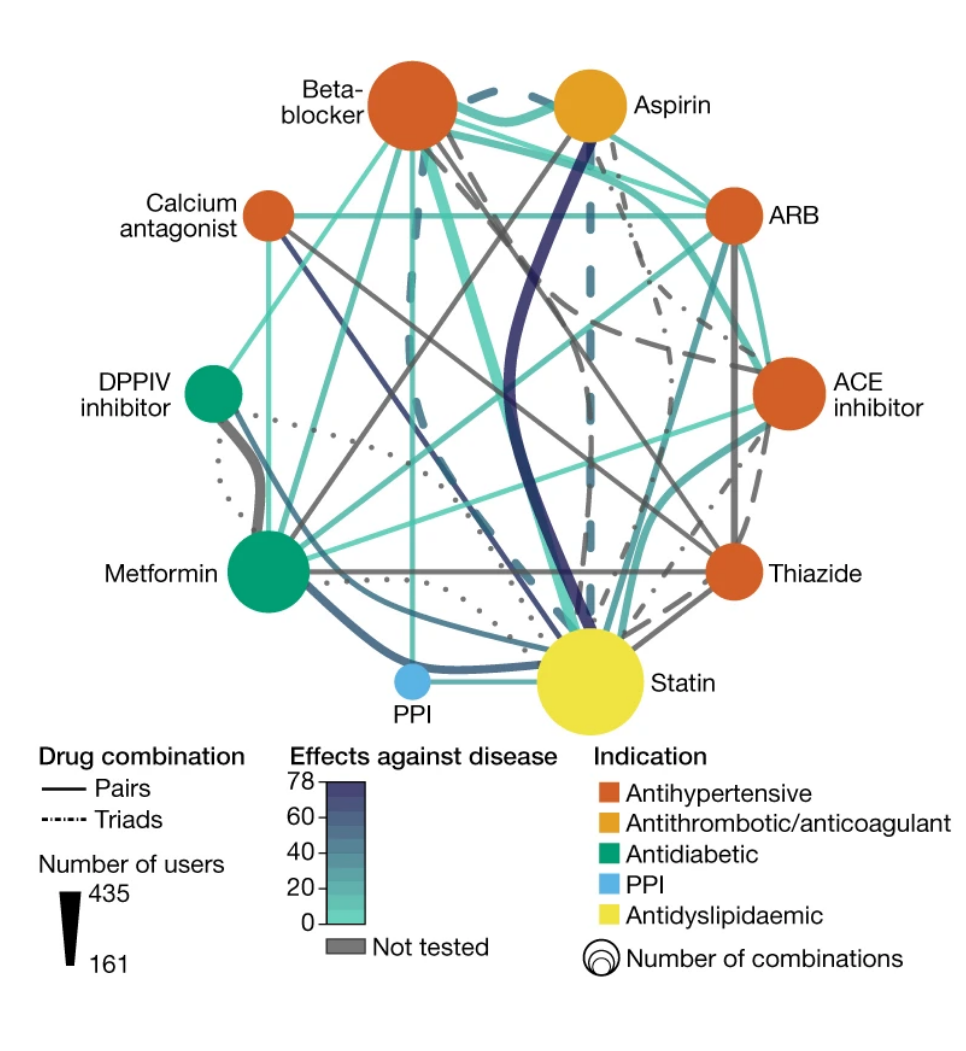
by Sofia K. Forslund, Rima Chakaroun, Maria Zimmermann-Kogadeeva, ..., Luis Pedro Coelho, ..., Peer Bork, The MetaCardis Consortium* in Nature
15. An open code pledge for the neuroscience community
by Cooper Smout, Dawn Liu Holford, Kelly Garner, ..., Sina Mansour L., Luis Pedro Coelho in Preprint
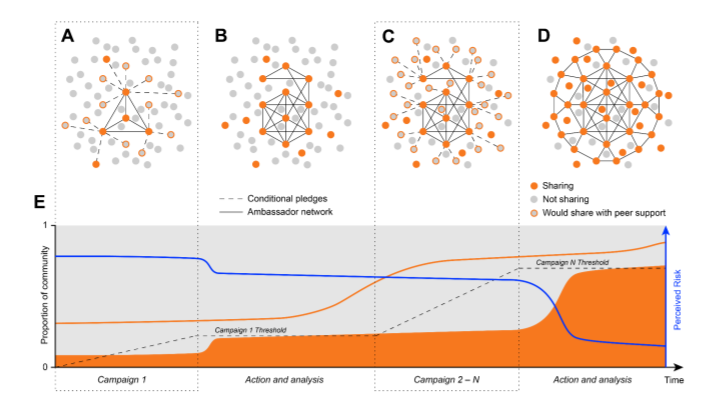
Conditional pledges can solve the collective action problem of code sharing (in neuroscience).
14. GUNC: detection of chimerism and contamination in prokaryotic genomes
by Askarbek Orakov, Anthony Fullam, Luis Pedro Coelho, Supriya Khedkar, Damian Szklarczyk, Daniel R. Mende, Thomas S. B. Schmidt, Peer Bork in Genome Biology
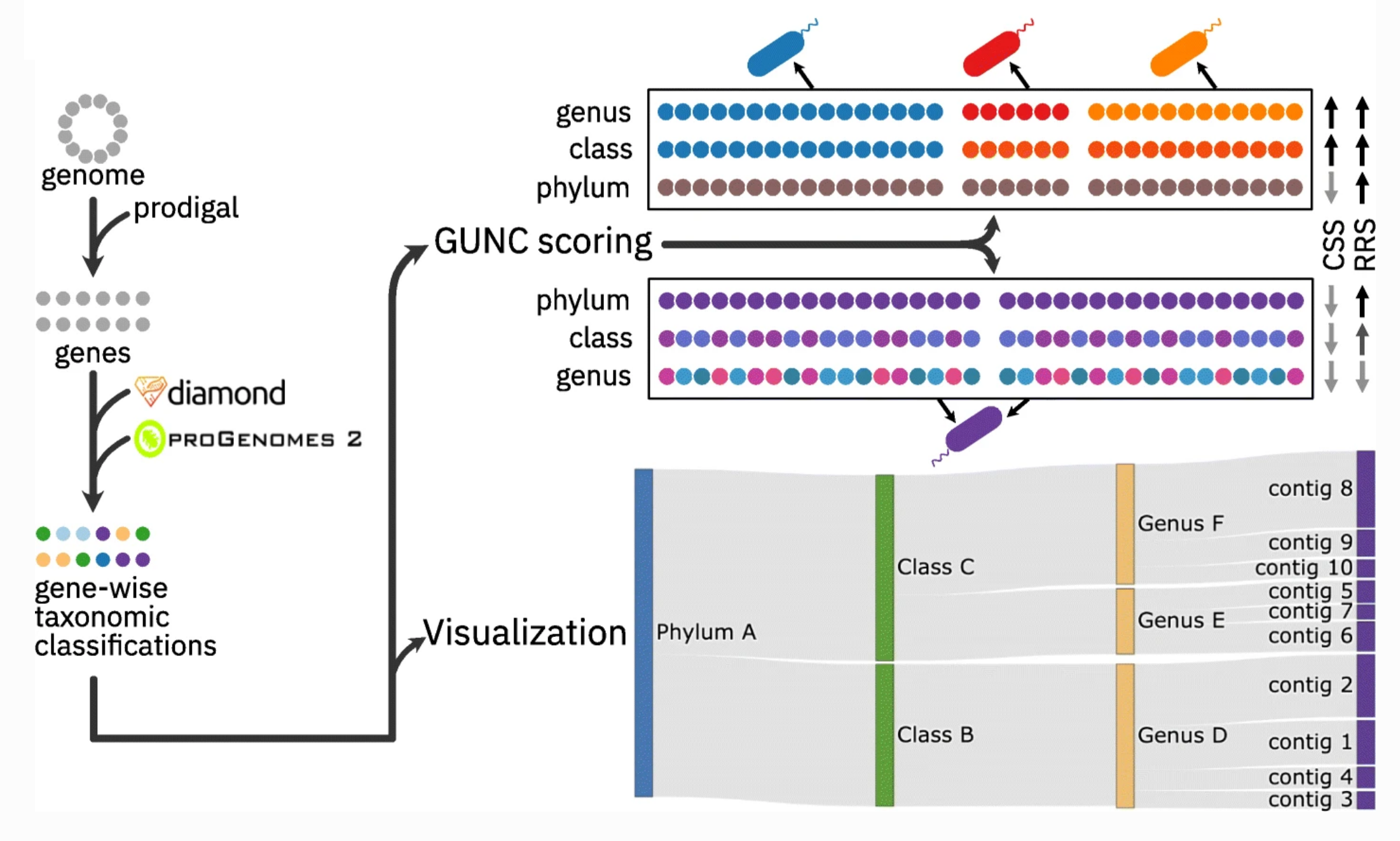
GUNC is a tool for finding chimeric genomes.
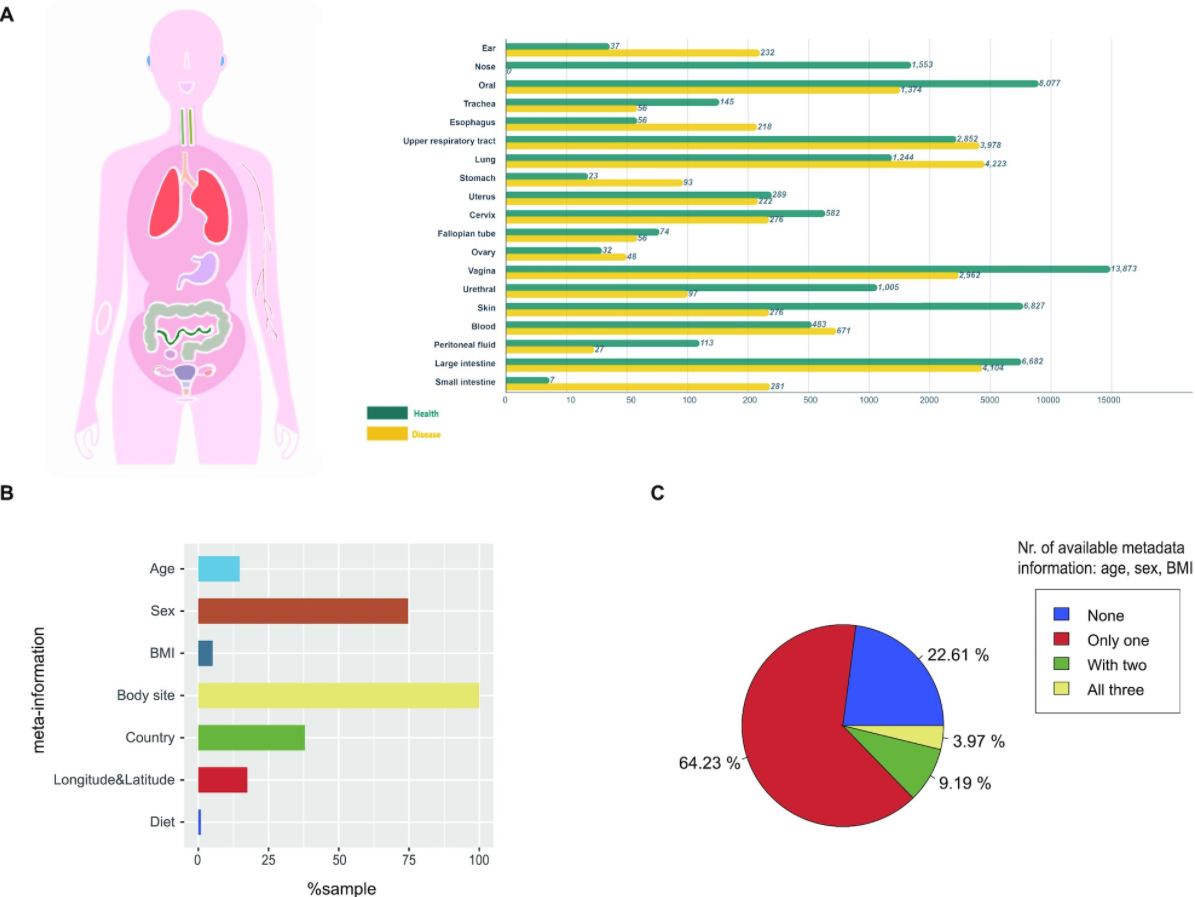
13. mBodyMap: a curated database for microbes across human body and their associations with health and diseases
by Hanbo Jin, Guoru Hu, Chuqing Sun, Yiqian Duan, Zhenmo Zhang, Zhi Liu, Xing-Ming Zhao, Wei-Hua Chen in Nucleic Acids Research
mBodyMap is a curated database for microbes across the human body and their associations with health and diseases.
DOI: 10.1093/nar/gkab973
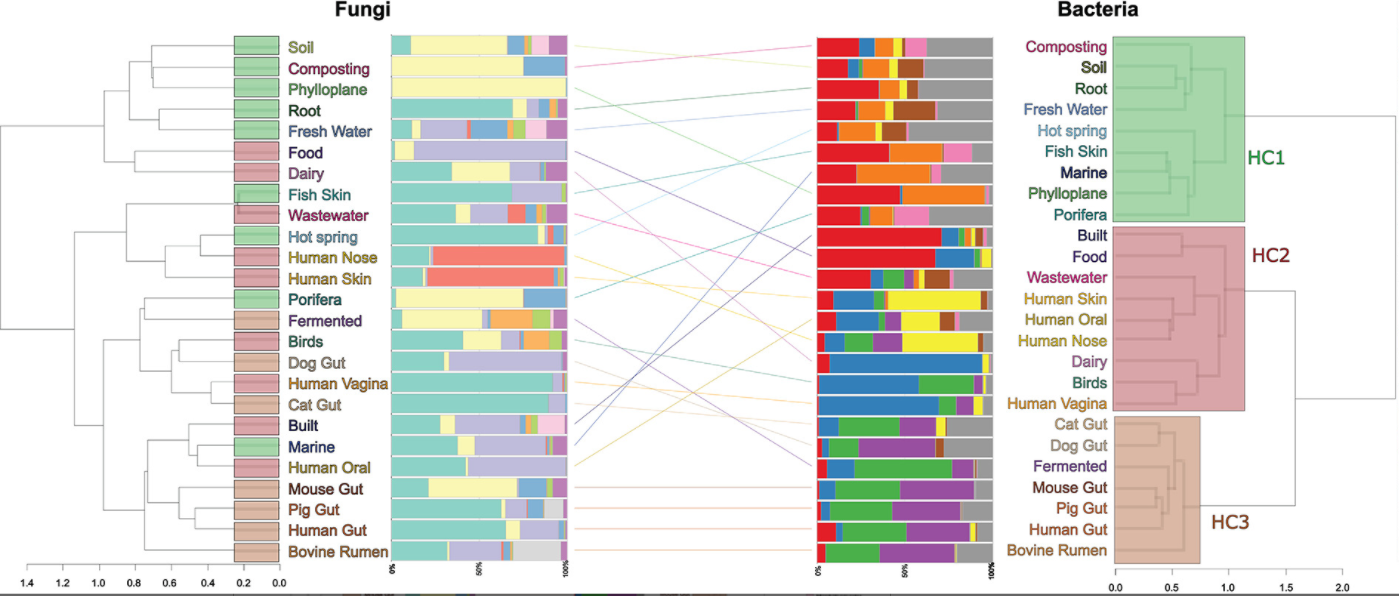
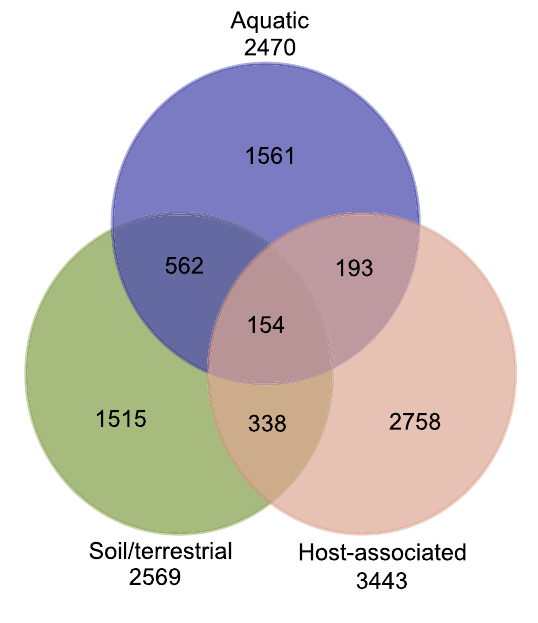
12. Metagenomic assessment of the global diversity and distribution of bacteria and fungi
by Mohammad Bahram, Tarquin Netherway, Clemence Frioux, Pamela Ferretti, Luis Pedro Coelho, Stefan Geisen, Peer Bork, Falk Hildebrand in Environmental Microbiology
Our results hint towards the strong role of environmental filtering in structuring cross-habitat bacterial but not fungal communities.
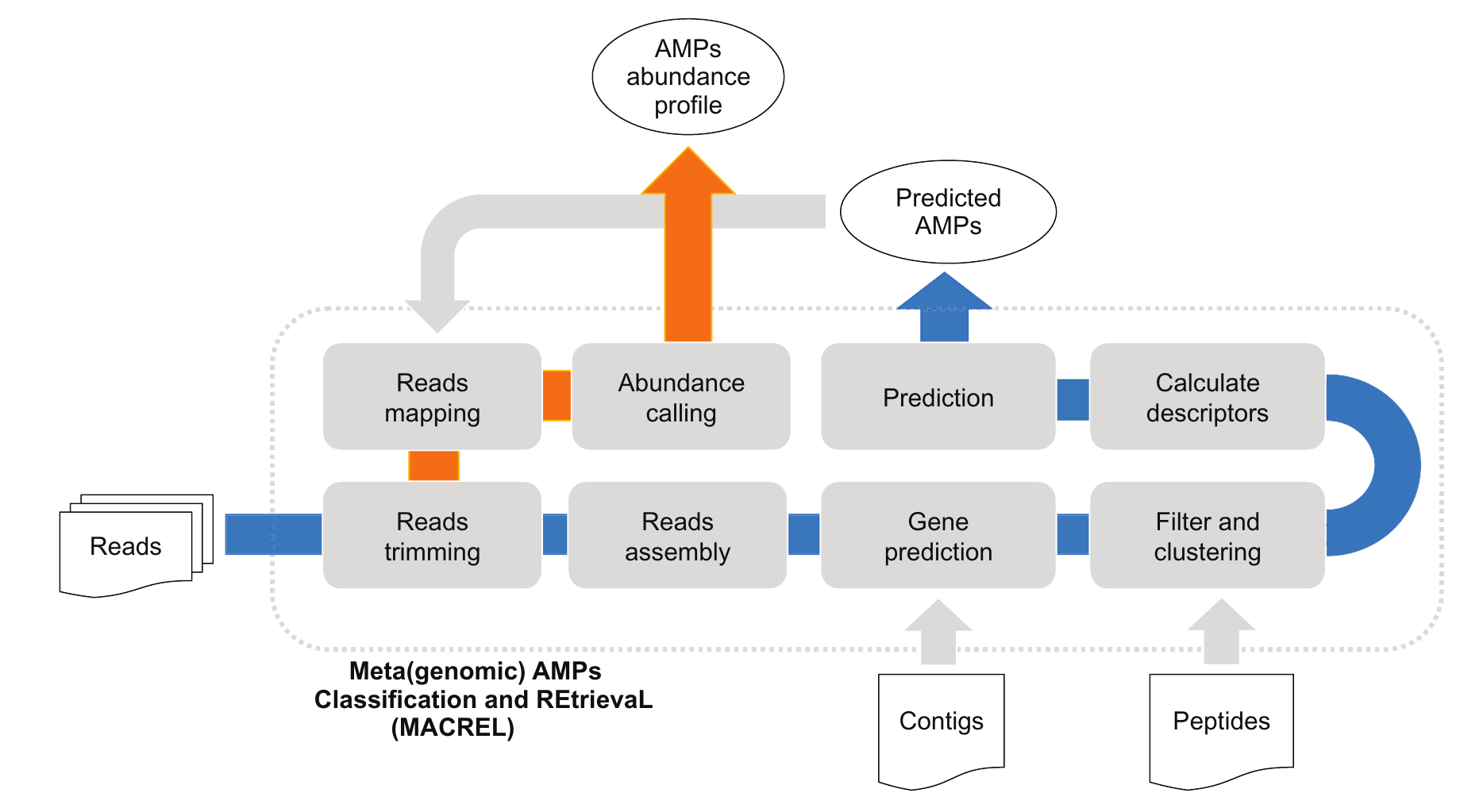
11. Macrel: antimicrobial peptide screening in genomes and metagenomes
by Célio Dias Santos Júnior, Shaojun Pan, Xing-Ming Zhao, Luis Pedro Coelho in PeerJ
Macrel is a tool for finding AMPs in (meta)genomes
DOI: 10.7717/peerj.10555
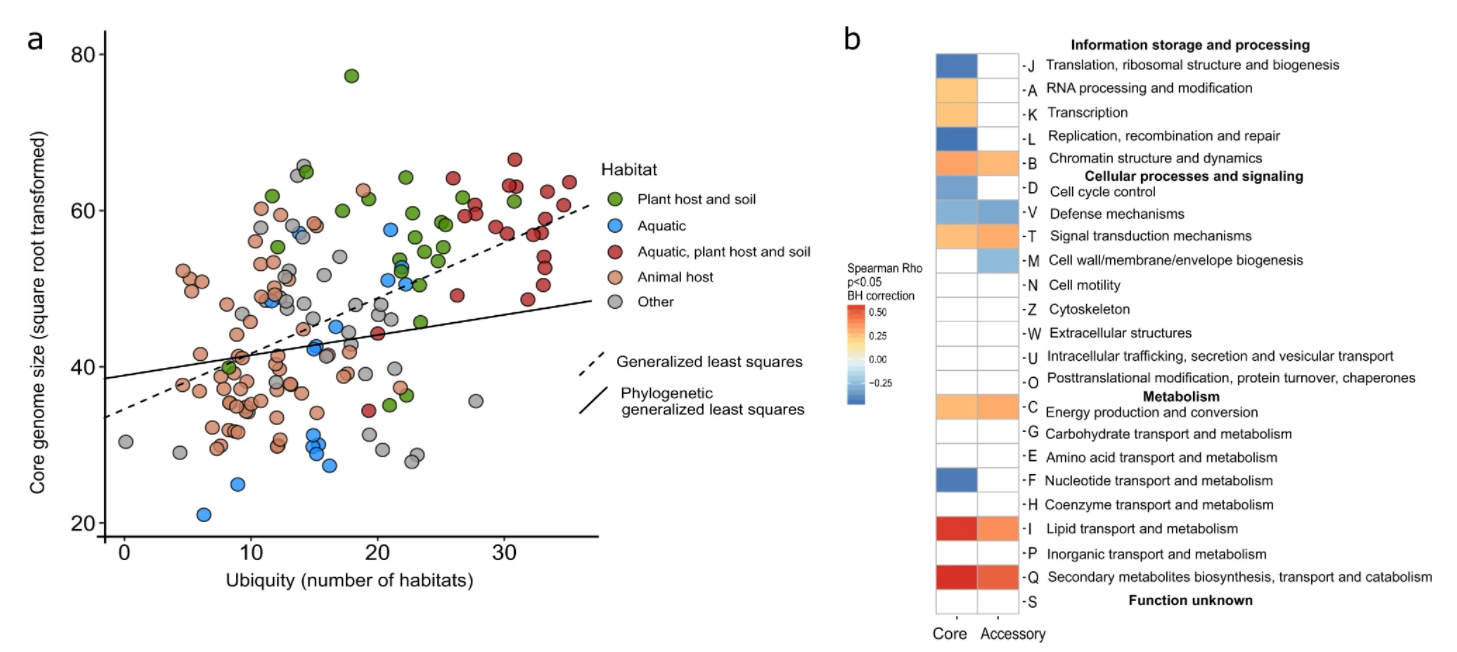
10. Disentangling the impact of environmental and phylogenetic constraints on prokaryotic within-species diversity
by Oleksandr M. Maistrenko, Daniel R. Mende, Mechthild Luetge, ..., Luis Pedro Coelho, ..., Shinichi Sunagawa, Peer Bork in The ISME Journal
We explored the global principles of pangenome evolution, quantified the influence of habitat, and phylogenetic inertia on the evolution of pangenomes and identified criteria governing species ubiquity and habitat specificity.
9. NG-meta-profiler: fast processing of metagenomes using NGLess, a domain-specific language
by Luis Pedro Coelho, Renato Alves, Paulo Monteiro, Jaime Huerta-Cepas, Ana Teresa Freitas, Peer Bork in Microbiome
NGLess is a tool for processing metagenomes. See more at https://ngless.embl.de
8. Microbial abundance, activity and population genomic profiling with mOTUs2
by Alessio Milanese, Daniel R Mende, Lucas Paoli, ..., Luis Pedro Coelho, ..., Georg Zeller, Shinichi Sunagawa in Nature Communications
We show that mOTUs, which are based on essential housekeeping genes, are demonstrably well-suited for quantification of basal transcriptional activity of community members.
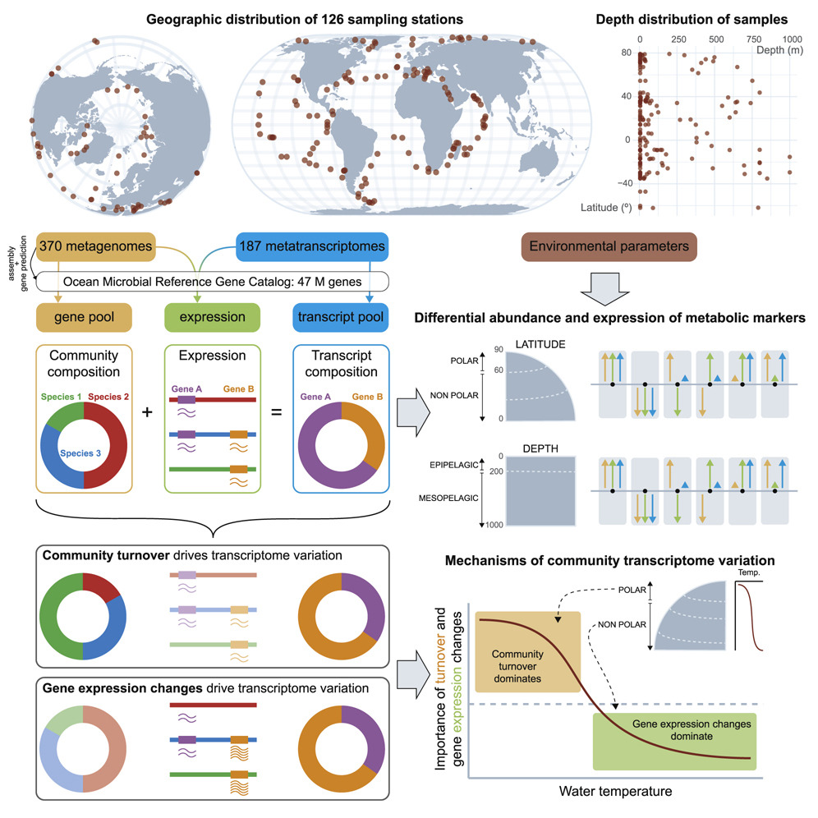
7. Gene Expression Changes and Community Turnover Differentially Shape the Global Ocean Metatranscriptome
by Guillem Salazar, Lucas Paoli, Adriana Alberti, ..., Luis Pedro Coelho, ..., Shinichi Sunagawa, Patrick Wincker in Cell
A New Meta-omics Resource for Global Ocean Microbiome Research
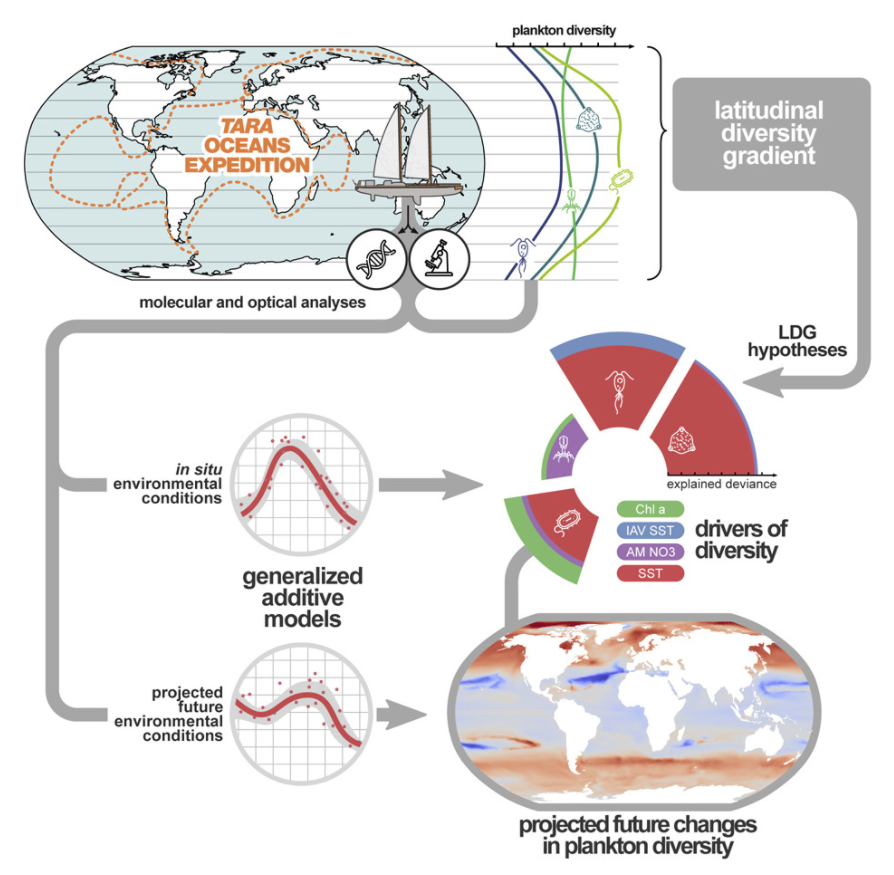
6. Global Trends in Marine Plankton Diversity across Kingdoms of Life
by Federico M. Ibarbalz, Nicolas Henry, Manoela C. Brandao, ..., Luis Pedro Coelho, ..., Shinichi Sunagawa, Patrick Wincker in Cell
Our results should therefore be seen as a baseline and a framework for testing new hypotheses about changes in diversity within the whole plankton community across the global ocean, identifying the most vulnerable areas, and to better appreciate and anticipate functional and socio-economic consequences.
5. proGenomes2: an improved database for accurate and consistent habitat, taxonomic and functional annotations of prokaryotic genomes
by Daniel R Mende, Ivica Letunic, Oleksandr M Maistrenko, ..., Luis Pedro Coelho, Peer Bork in Nucleic Acids Research
Progenomes2 is a collection of well-annotated, high quality prokaryotic isolate genomes.
DOI: 10.1093/nar/gkz1002
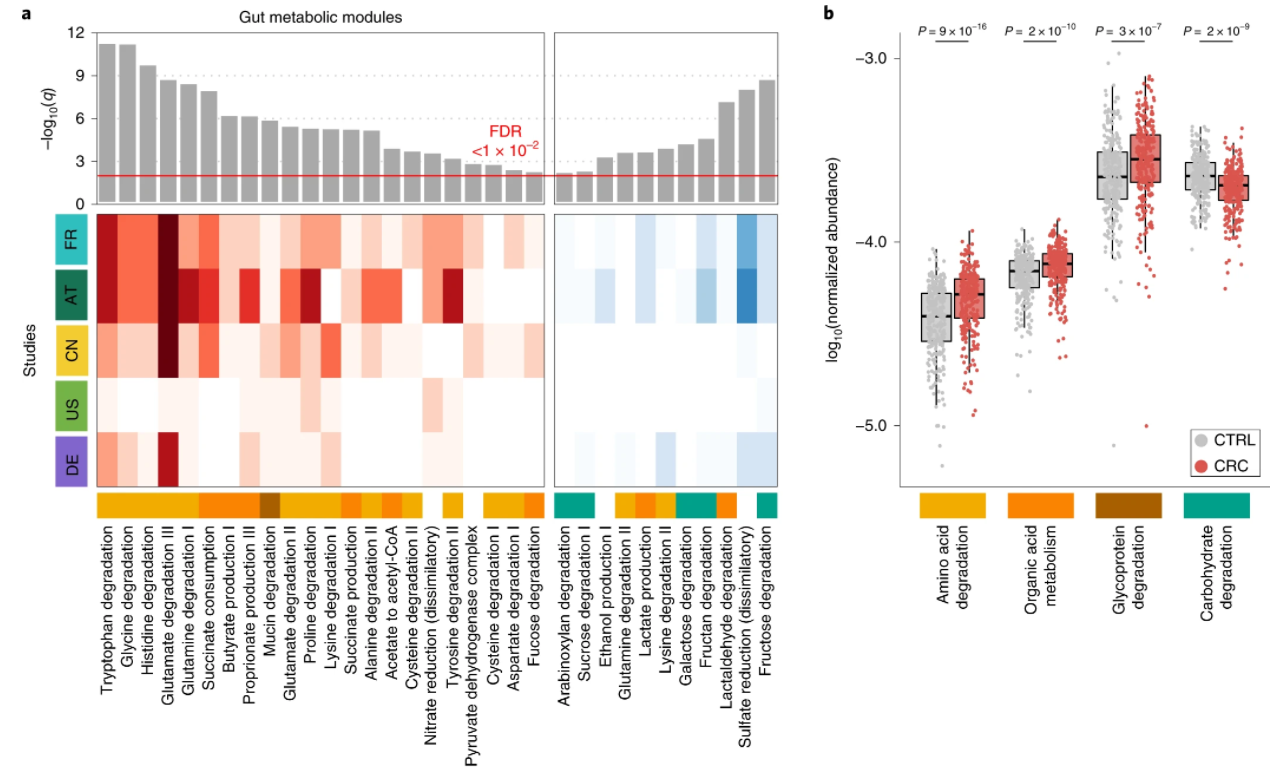
4. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer
by Jakob Wirbel, Paul Theodor Pyl, Ece Kartal, ..., Luis Pedro Coelho, ..., Peer Bork, Georg Zeller in Nature Medicine
Through extensive validations, this meta-analysis firmly establishes globally generalizable, predictive taxonomic and functional microbiome CRC signatures as a basis for future diagnostics.
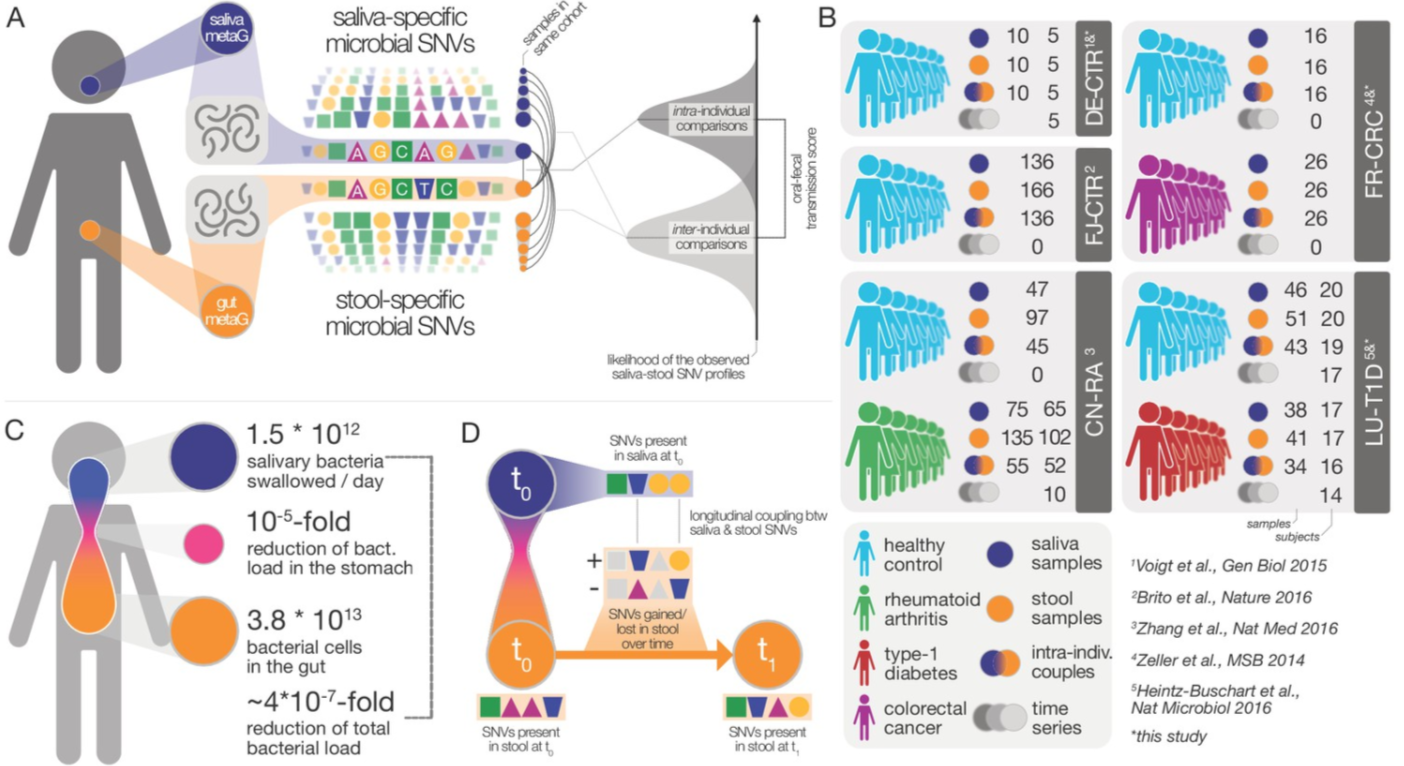
3. Extensive transmission of microbes along the gastrointestinal tract
by Thomas SB Schmidt, Matthew R Hayward, Luis Pedro Coelho, ..., Paul Wilmes, Peer Bork in eLife
We found that transmission to, and subsequent colonization of, the large intestine by oral microbes is common and extensive among healthy individuals.
DOI: 10.7554/elife.42693
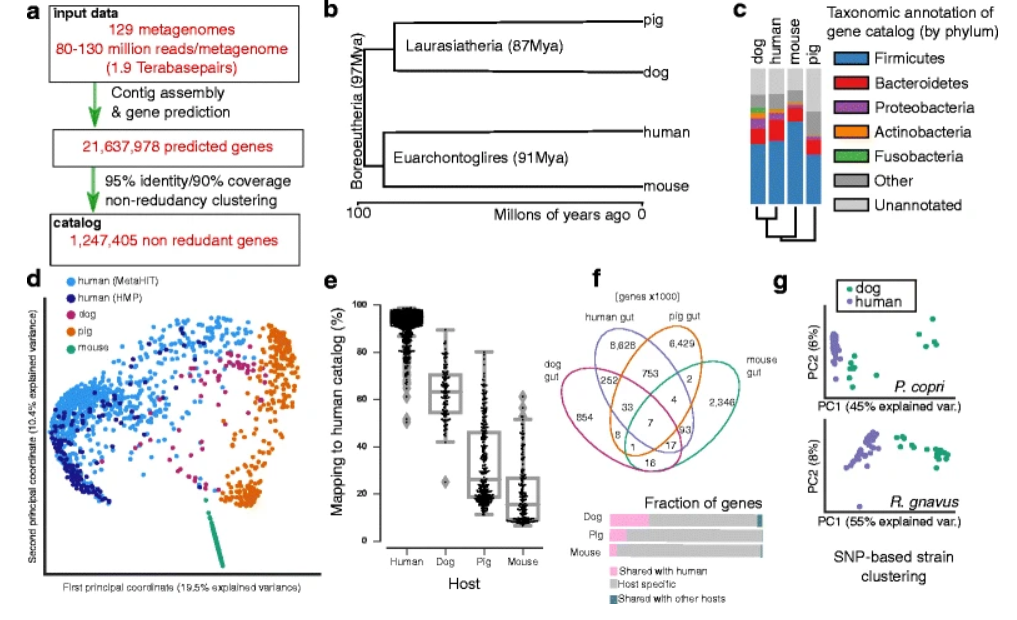
2. Similarity of the dog and human gut microbiomes in gene content and response to diet
by Luis Pedro Coelho, Jens Roat Kultima, Paul Igor Costea, ..., Qinghong Li, Peer Bork in Microbiome
We conclude that findings in dogs may be predictive of human microbiome results. In particular, a novel finding is that overweight or obese dogs experience larger compositional shifts than lean dogs in response to a high-protein diet.
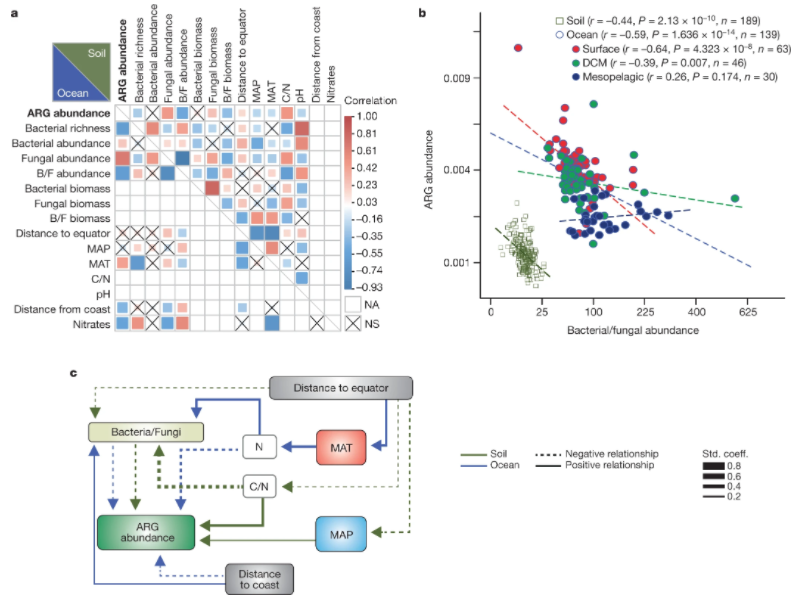
1. Structure and function of the global topsoil microbiome
by Mohammad Bahram, Falk Hildebrand, Sofia K. Forslund, ..., Luis Pedro Coelho, ..., Leho Tedersoo, Peer Bork in Nature
Our results suggest that both competition and environmental filtering affect the abundance, composition and encoded gene functions of bacterial and fungal communities, indicating that the relative contributions of these microorganisms to global nutrient cycling varies spatially.
Copyright (c) 2018–2025. Luis Pedro Coelho and other group members. All rights reserved.