BDB-Lab members involved
ResFinderFG v2.0: a database of antibiotic resistance genes obtained by functional metagenomics
ResFinderFG v2.0: a database of antibiotic resistance genes obtained by functional metagenomics
by Rémi Gschwind, Svetlana Ugarcina Perovic, Maja Weiss, Marie Petitjean, Julie Lao, Luis Pedro Coelho, Etienne Ruppé
Abstract
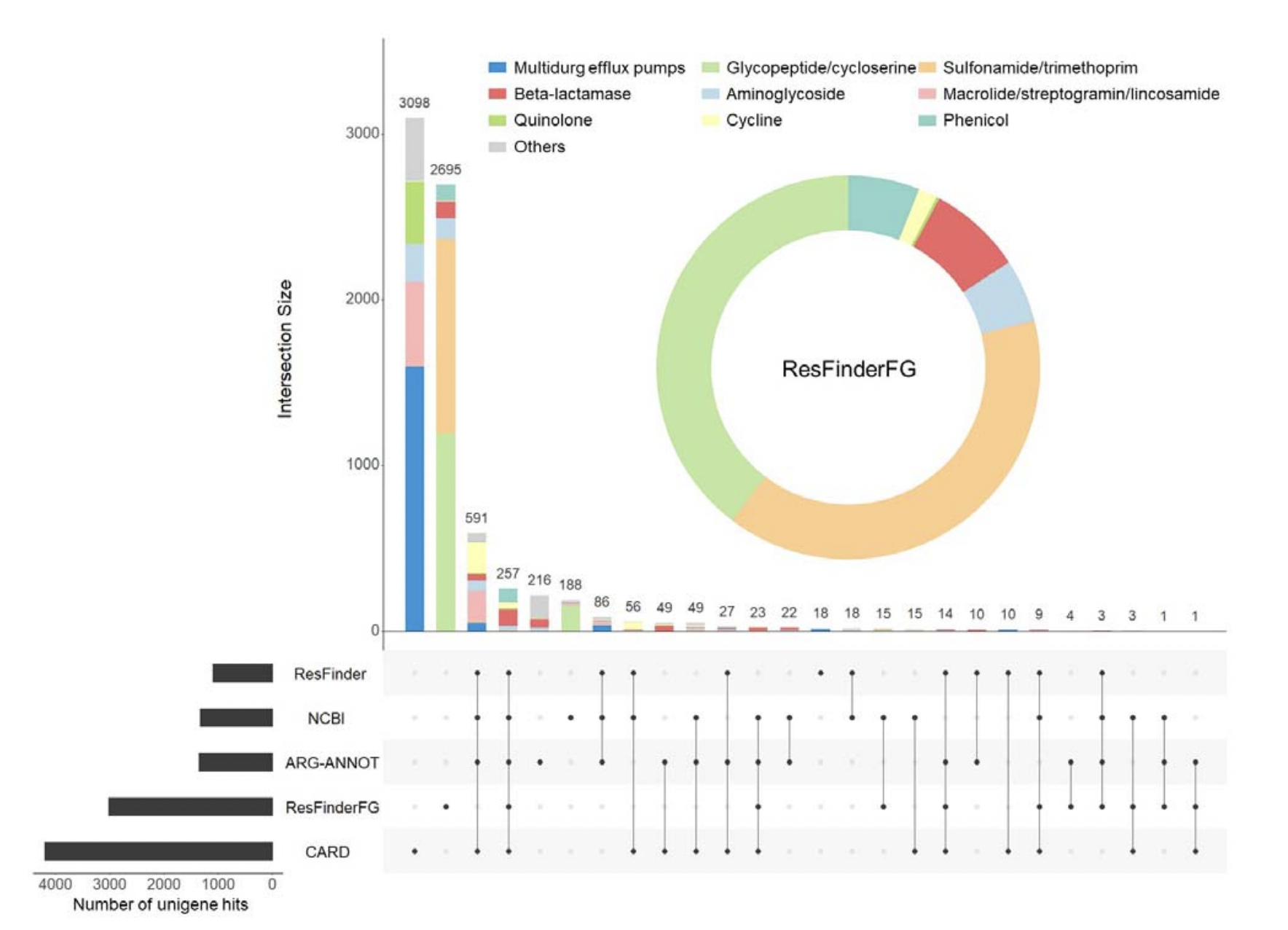
In 2016, the ResFinderFG v1.0 database was created to collect ARGs from functional metagenomics studies. Here, we present the database second version, ResFinderFG v2.0. Fifty publications were considered, for a total of 23’764 ARGs identified from different environments. After deduplication, annotation and curation, 3’913 ARGs were included. New ARGs included are mainly glycopeptides/cycloserine or beta-lactams resistance genes identified mostly in human-associated samples. Results of GMGC gene subcatalogs annotation showed that ResFinderFG v2.0 detected comparable or higher ARG numbers than those detected with other databases. Most of the unigene hits obtained were database-specific and ResFinderFG v2.0 specific unigene hits included among others: glycopeptides/cycloserine, sulofnamides/trimethoprim resistance genes and beta-lactamases encoding genes. ResFinderFG v2.0 can be used to identify ARGs differing from those found in conventional databases and therefore improve the description of resistomes.
Full text: https://doi.org/10.1093/nar/gkad384
Copyright (c) 2018–2024. Luis Pedro Coelho and other group members. All rights reserved.
