Most recent BDB-Lab papers
BDB-Lab Links
Major Projects
Other Links
Lab Members
Twitter feed
Tweets by BigDataBiologyAzuC: A non-conventional AMP candidate
by Tobi Olanipekun, Celio Dias Santos Junior, Luis Pedro Coelho.
AzuC was predicted as an AMP
Similar to what we had done for hg4, we used macrel to predict potential AMPs in hundreds of publicly available metagenomes. When cataloging these peptides, some of them were homologous to AzuC, a protein of unknown function, suggesting to us that AzuC could be an AMP. While we are still awaiting further confirmatory work, this is a tempting hypothesis.
AzuC, despite its gene being widely studied, still remains with an unknown biological function. This is due to its small size which makes purification and characterization difficult. Recently, Guyet et al. (2018) found azuC gene expression responding linearly to pressures between 0 MPa to 1 MPa. The same authors still conclude:
It is tempting to speculate that AzuC was upregulated in response to mild elevated pressure to maintain membrane stability just like Csps in higher pressure treatments.
Other authors found azuC over expressed under other stress conditions such as low pH, heat shock and oxidative stress, although it was regulated under low oxygen levels. AzuC, and other stress induced small membrane proteins, could stabilize the membrane by the modulation of other transmembrane proteins and also by interacting with the inner membrane changing its permeability (Hemm et al. 2010). This interaction with membranes, changing their permeability, is a key feature of many AMP classes.
A series of in silico tests were carried out on these sequences so as to screen for antimicrobial features or activities of the predicted peptides, we got interesting results that can serve as evidences supporting our speculation of AzuC being an AMP-like protein, furthermore, we went on to document some of its peptide features and also to model it. These results are the major scope of this write-up.
Analyzing the peptide
We eliminated the identical sequences that matched to AzuC by using CD-HIT with sequence identity cut-off of 100% and overlap of the shorter sequence by 90%. We initially got 948 sequences clustered into 12 clusters. a href="http://skylign.org/"Skylign was used to create logos (below) of the 60bp upstream and downstream of the start and stop codons of each gene. As in HG4, there is a ribosome binding site with a 7bp spacer in the upstream region, and on the downstream region, a stop codon. Thus there is a complete ORF that can be transcribed, including a terminator at the downstream portion.
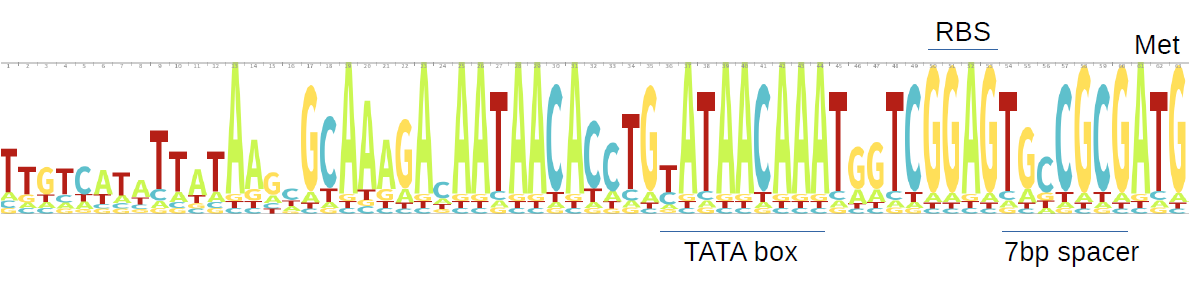
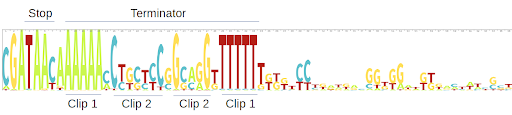
After an evolutionary analysis of the azuC genes using a href="http://selecton.tau.ac.il/"Selecton, we found evidence of purifying selection. This was also supported by a negative Tajima’s D (-0.74), thus suggesting amino acid conservation. The found genes were very close to each other as shown in the phylogeny, suggesting they are closely related.

Features of a typical AMP could be verified in AzuC peptides, such as the amphiphilic profile, with the charged amino acids preferentially distributed on one side of the helical wheel. The other face of the wheel is occupied mostly by hydrophobic residues, which can also be seen in its hydrophobicity profile, with alternating sections of polar and hydrophobic regions. This alternancy is typical for AMPs, facilitating their insertion in membranes.

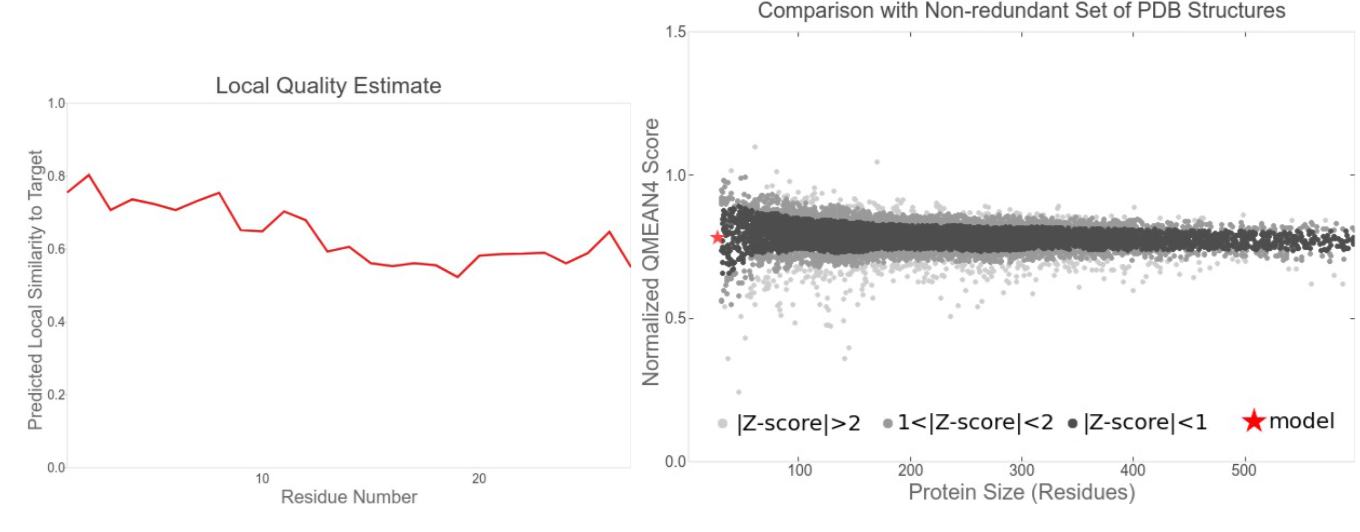
The structure of the AzuC protein was predicted using SwissModel, and the overall quality measured by the QMEAN (0.25) was extremely close to an experimentally obtained model, also corroborated by the Z-value (<1). Altogether, these parameters suggest our model as feasible to infer at least some preliminary conclusions about azuC.
AzuC peptide model is shown in blue (below), and is clearly formed mostly by an alpha-helix, as most other AMPs (Zhang and Gallo, 2016). Like bacteriocins, it has an arm, with charged amino acids that can anchor in the membrane and stabilize the peptide/membrane complex. After tests of melting, the structure showed to be thermodynamically stable with an RMSD of the structures of 3.0, and a maximum energy of -300 kJ/mol.

AzuC: an unusual AMP candidate
As we found azuC in human gut samples, we predicted its half life in intestine-like environments using the HLP WebServer. The predict half-life of azuC in human guts is 0.065s; in other words, it has low stability (half life <0.1s - Sharma et al., 2014). Although no signal peptides were predicted, azuC still has some AMP targets predicted using the special prediction tool from DBAASP server. AzuC was predicted to be active against Klebsiella pneumonia (0.85 PPV) and Bacillus subtilis (0.81 PPV), the latter a well-known spore-forming bacteria. The interest in the control of both bacteria and also the niche of both species in relation to E. coli makes azuC a good candidate for testing in vitro against these strains to check its effectiveness, although no actual evidence of its potency could be predicted.
Considering the origin of azuC genes, mostly from E. coli and other Gram-negative microorganisms, its pattern of expression under stress conditions, and the clear interaction with membranes already documented and circumstantially linked to a modulation in plasticity, leads us to a funny line of thought:
- azuC kept its properties and still is capable of interacting with membranes
- its original function was killing competitor strains
- no signal peptide was found and expression is up-regulated under stress
Conclusions and perspectives
We need to understand that besides low stability, azuC lacks a signal peptide (cannot be secreted by conventional pathways) and is produced mostly in stress conditions. These pieces of evidence can lead us to conclude that AMPs can have other functions than killing microorganisms. Specifically, we are tempted to speculate that E. coli (the bacteria from which azuC gene was identified) is using the AMP features to modulate the plasticity of its own membrane and therefore modulate the cell shape and volume. This circumstantial evidence, if further corroborated in other genes, can show that AMPs could evolve into membrane proteins. Similar findings were recently reported by Garrido et al. (2020) where they show the emergence of targeting peptides from AMPs for the evolution of endosymbiotic protein targeting systems.
References
Hemm MR, Paul BJ, Miranda-Rios J, Zhang AX, Soltanzad N, Storz G. Small Stress Response Proteins in Escherichia coli: Proteins Missed by Classical Proteomic Studies. J Bacteriol. 2010;192(1):46–58. WOS:000272636600006. pmid:19734316
Garrido, C.; Caspari, O.D.; Choquet, Y.; Wollman, F.-A.; Lafontaine, I. Evidence Supporting an Antimicrobial Origin of Targeting Peptides to Endosymbiotic Organelles. Cells 2020, 9, 1795.
Zhang LJ, Gallo RL. Antimicrobial peptides. Curr Biol. 2016;26(1):R14-R19. doi:10.1016/j.cub.2015.11.017
Sharma A, Singla D, Rashid M, Raghava GP. Designing of peptides with desired half-life in intestine-like environment. BMC Bioinformatics. 2014;15(1):282. Published 2014 Aug 20. doi:10.1186/1471-2105-15-282
Copyright (c) 2018–2025. Luis Pedro Coelho and other group members. All rights reserved.