BDB-Lab members most involved
Small Proteins of the Global Microbiome
Small proteins (or small open reading frames, smORFs) are often ignored computationally because of the technical difficulties they present, even though recent studies indicated that small proteins have important functions in cellular biology, such as the regulation of amino acid metabolism and antimicrobial activity.
At the BDB-Lab, we have several sub-projects attempting to understand the role they play in prokaryotic biology. One particular subset of small proteins that we started to investigate are AMPs (anti-microbial peptides). These molecules are used by different microbes to modulate their growth as colonies or even increase their fitness in the inter- and intra-species competition. Currently, AMPs have a high potential to treat persistent diseases, with some of them already being used as supplements or food additives (e.g., nisin).
Macrel
The recent growth in the availability of genomes and metagenomes provides an opportunity for in silico prediction of novel AMP molecules. However, AMPs are too small for standard gene prospection methods.
Macrel (for metagenomic AMP classification and retrieval) is an end-to-end pipeline for the prospection of high-quality AMP candidates from (meta)genomes. Its classifiers perform similarly to the state-of-the-art in the prediction of both antimicrobial and hemolytic activity of peptides. However, Macrel has enhanced precision, recovering high-quality AMP candidates using real data.
Macrel is implemented in Python and is available as open source.
Macrel Links
- Macrel webserver
- Macrel repository
- Manuscript: Santos-Júnior CD, Pan S, Zhao XM, Coelho LP. Macrel: antimicrobial peptide screening in genomes and metagenomes. PeerJ. 2020 Dec 18;8:e10555. doi: 10.7717/peerj.10555. PMID: 33384902; PMCID: PMC7751412.
AMPSphere - the world wide survey for AMPs
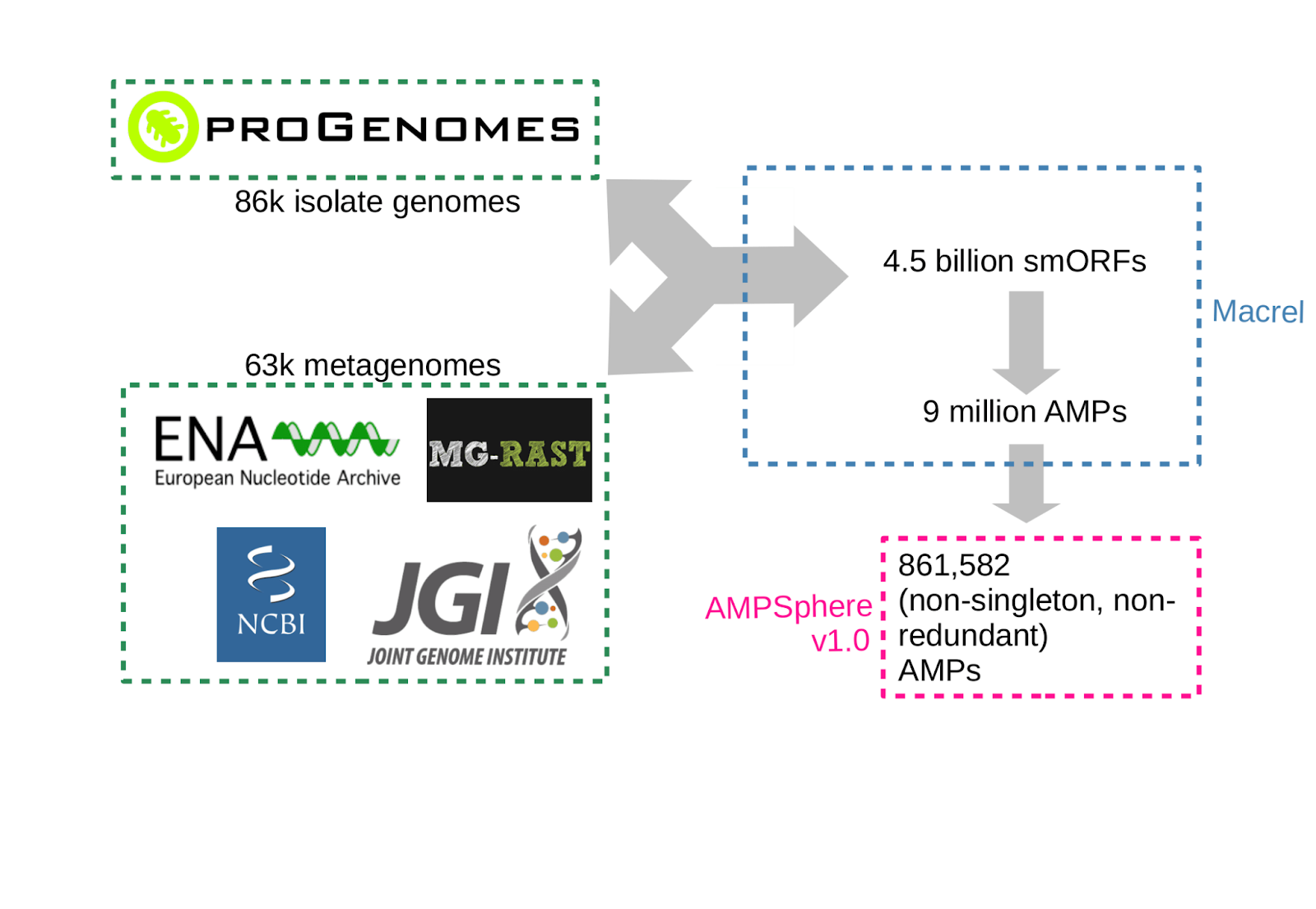
AMPSphere (the world wide survey for AMPs) aims to sample and catalog the antimicrobial peptides (AMPs) of the global microbiome. AMPs are operationally defined as proteins containing 10-100 residues, that are predicted to disrupt microbial growth. Applying macrel to metagenomes and isolate genomes, and eliminating redundancy, we obtained a catalog of ca. 1 million AMPs.
We are currently attempting to better understand the evolutionary origin of these genes. One route for the creation of smORFs is that they derive from larger genes, which were disrupted forming smaller proteins, becoming independent biological entities.
This project is conducted in collaboration with the Bork Group at EMBL and the Huerta-Cepas group at CPGB.
Working directly on the AMPs, Dr. Celio Dias Santos Junior, a biotechnologist with expertise in high-throughput sequencing, metagenomics and transcriptomics to characterize environmental microbiomes. He is a postdoctoral researcher in BDB since 2019, and is interested in the distribution, diversity and dynamics of global antimicrobial peptides (work presented at the World Microbe Forum 2021).
Celio has been mentoring undergraduate students developing side-projects related to BDB main research interests, including students signed within the NSURP (The National Summer Undergraduate Research Project) and our BDB internship program.
AMPSphere Links
- Permalink to the AMPSphere resource
- AMPSphere resource website:
- http://ampsphere.big-data-biology.org/ (currently still a WIP)
GMSC - Global Microbial Small ORFs Catalog
While the AMPSphere focuses on AMPs, whose function can be predicted with some confidence, that still ignores the vast majority of smORFs. Therefore, we are constructing a global microbial smORFs catalog (GMSC, by analogy with the GMGC), which will provide a complete annotated resource for smORFs in the global microbiome.
So far, we predicted 4.5 billion smORFs and clustered them at 100%, 90%, and 50% amino acid sequence identity. A current focus is to further filter these large datasets to obtain a high-quality subset.
Yiqian Duan is a bioinformatician with expertise in high-throughput sequencing to characterize environmental microbiomes. She is a Ph.D. candidate in BDB since 2020, and is interested in the distribution, diversity and dynamics of small ORFs. Currently, Yiqian and Dr. Celio Dias Santos Junior are conducting the investigations on the collaborative work involving small ORFs.
This project is conducted in collaboration with the Bork Group at EMBL and the Huerta-Cepas group at CPGB.
Side Projects
Amy Houseman developed a directed analysis of a cryptic peptide candidate in human-associated samples. It was called HG4 and was found in Prevotella This was the initial hint for us that this could be an important mechanism.
More recently, Anna Vines has showed us a more holistic view on the cryptides by searching for matches to the AMPSphere on the whole of progenomes2.
Tobi G. Olanipekun analyzed an AMP candidate called AzuC, a gene that was previously described in Escherichia coli and has so far no associated known function. His work is described in a blogpost.
Copyright (c) 2018–2024. Luis Pedro Coelho and other group members. All rights reserved.

